Physiological, Biochemical, and Molecular-Genetic Events Governing the Recognition and Resolution of RNA/DNA Hybrids
- Robert J. Crouch,
PhD, Head, Section on Formation of RNA - Susana M. Cerritelli, PhD, Staff Scientist
- Kiran Sakhuja, MS, MSc, Biologist
- Andrea Mota, BS, Postbaccalaureate Fellow
- Alexander Robinson, BS, Postbaccalaureate Fellow

Ribonucleases H (RNases H) are considered essential enzymes in multicellular organisms, thereby placing the genes that encode the enzymes in the housekeeping category. Damaged DNA is a leading cause of many human diseases and disorders. We study the formation and resolution of RNA/DNA hybrids, which occur during DNA replication and RNA transcription. Such hybrid molecules may lead to increased DNA damage, but may also play critical roles in normal cellular processes. We are interested in how RNA/DNA hybrids are resolved and in the role that RNases H play in their elimination. Two classes of RNases H, Class I and Class II, are present in most organisms.
Patients with mutations in the RNASEH1 gene exhibit typical mitochondrial myopathy symptoms (neuromuscular disorder). We were the first to show that RNase H1 is essential for the maintenance of mitochondrial DNA. More than 1,000 proteins are targeted to mitochondria, many of which, when mutated, are known causes of mitochondrial myopathies. Mice deleted for the Rnaseh1 gene arrest embryonic development at day 8.5 because of failure to amplify mitochondrial DNA.
Aicardi-Goutières syndrome (AGS), a severe neurological disorder with symptoms appearing at or soon after birth, can be caused by defective human RNase H2. As many as 38 Mendelian genotypes may result in a type I interferonopathy, including mutations in each of the genes encoding the subunits of the heterotrimeric RNase H2, the hallmark of which is activation of the innate immune response.
RNA/DNA hybrids, ribonucleotides embedded in DNA: overlapping and disparate resolution by RNases H and the RNA exosome
Ribonucleases H in all kingdoms
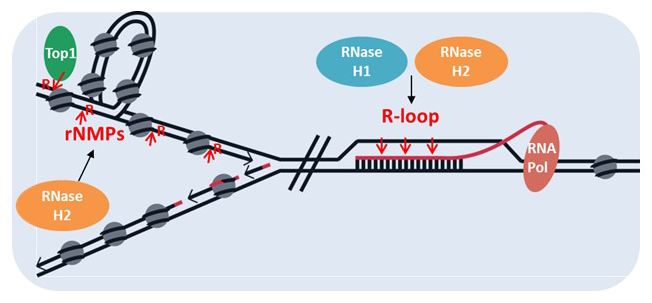
Ribonucleases H are present in all kingdoms. They were originally described and purified based on their ability to cleave the RNA of RNA/DNA hybrids, a duplex with one RNA strand complementary to a DNA strand. Substrates for RNases H, at least when used in biochemical analysis, (1) need to be stable, and (2) the minimum length of RNA is four rNMPs (ribonucleoside monophosphates) for type I enzymes, whereas a single rNMP embedded in duplex DNA is incised by type II RNases H. Single rNMPs are incorporated by DNA polymerases with frequencies dependent on the polymerase. The three replicative DNA polymerases have different affinities for rNMPs, resulting in about one rNMP per 12,000 dNMPs (deoxyribonucleoside monophosphates) incorporated. RNA/DNA hybrids are formed during transcription when, after the nascent RNA exits the RNA polymerase, it invades the underwound trailing DNA, displacing the non-template strand, forming an R-loop (three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA). In the bacterium Escherichia coli, two RNases H are present, one able to incise at single rNMPs and the other very active on RNA/DNA hybrids. E. coli cells are viable when either one or both RNases H are deleted. Eukaryotic RNases H have one RNase H that resembles type I RNase H (H1) and a second RNase H (H2), which can recognize single rNMPs in DNA as well as, sharing with H1, the hydrolysis of RNA of RNA/DNA hybrids. In the yeast Saccharomyces cerevisiae, H1 and H2 are not required for viability, either in single or double deletion. Mouse and human RNases H are both essential.
RNases H in E. coli, S. cerevisiae, and mouse
E. coli cells with loss of RNase HI are unable to grow in the absence of the recombination enzyme RecBCD (a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA), a complex that is dispensable when RNase HI is present. This type of paired lethality, termed synthetic lethality, can aid in deciphering the various substrates the pair of enzymes have or the pathways they share. We constructed an E. coli strain in which we could conditionally knockout the rnhA gene in a RecBCD–defective background. We selected 30 very rare survivors following knockout of the rnhA gene, on whose genomes we performed long read sequencing. The best description of the results is chaos! E. coli, like most bacteria, have transposons and endogenous viruses called prophages in their genomes. Most of the strains sequenced have transposons present in places other than their original location. One strain is noteworthy with eight copies of a region containing 16 complete genes and two partial genes. INSAB, one of the E. coli transposons, was inserted in this region, an insertion that is known to amplify adjacent genes by recombination using the recA pathway. Amongst all the noise resulting from the chaos, all survivors had a mutation in the rnlA gene. The RnlA protein is a ribonuclease that is inactive under normal conditions by interaction with the RnlB protein. Viral infection frees the RnlA protein to degrade endogenous RNA. The bacteriophage T4 counters this activation by expressing an inhibitor of RnlA, allowing production of virus particles. Our results suggest that active RnlA is detrimental to the rnhA-recBCD–deleted cells and must be mutated for survival. We found multiple forms of rnlA inactivation in our mutants, such as frameshifts creating stop codons, insertion of one of two retrotransposons, deletion of several genes flanking the rnlA gene, and complete loss of the prophage in which the rnlA resides. It has been reported that RNase HI and RnlA physically interact, and we also have observed this interaction using AlphaFold, an AI program that predicts protein structure. RNA/DNA hybrids can be flanked by single-stranded RNA. A complex between RNase HI and RnlA could be a means to completely remove the remaining transcript. Conversely, the RnlA nuclease could use the RNA/DNA hybrid RNase H activity when hybrids are encountered by RnlA.
The RNA exosome and RNases H cooperate to suppress R-loop–mediated genome instability.
There are numerous cellular factors that prevent and/or process R-loop formation and stabilization. In addition to RNases H, RNA/DNA helicases can resolve hybrids and eliminate R-loops. Cellular processes, such as mRNA packaging and export, avoid hybrid formation. The RNA exosome is a major 3′-5′ RNA–degradation, multi-subunit complex in eukaryotes, which degrades cryptic and defective transcripts, preventing their engagement with DNA and the formation of R-loops. In collaboration with Aziz El Hage and David Tollervey, we are studying the interactions between the RNA exosome and RNases H in suppressing R-loop–mediated genome instability.
Using the yeast S. cerevisiae, we found that cells defective for both RNase H and RNA exosome activities are hypersensitive to the drug hydroxyurea, which induces replicative stress by depleting the cellular dNTP (deoxynucleotide triphosphate) supply. Replicative stress in these conditions can be eliminated by expressing the RNase H2-RED variant, which processes R-loops, indicating that the defects observed are caused by R-loop accumulation in the absence of fully active RNases H and RNA exosome complexes. Overexpression of RNase H1 partially suppressed the growth defects of exosome-deficient mutants, suggesting that some of the problems in these cells are caused by harmful R-loops.
We are employing a genetic system that correlates R-loop accumulation with homologous recombination to further elucidate the interplay between RNases H and the RNA exosome in R-loop processing. In this system, yeast strains contain a fragment of the mouse switch Mu sequence (from the IgH locus) in an orientation that either facilitates or prevents R-loop formation. We found that RNAs with the fragment in both orientations accumulated in exosome-deficient mutants, indicating that these RNAs are targeted for degradation by the exosome. Moreover, strains with the fragment in the orientation that favors R-loop formation showed increased accumulation of RNA/DNA hybrids and higher recombination rates when both the exosome and RNases H were defective, compared with cells lacking only RNases H. We conclude that RNase H and the exosome pathways converge to promote genome stability by suppressing the harmful effects of R-loops.
More recently, we began genome-wide mapping of R-loops in RNA exosome–defective cells, which lack RNase H1 activity. We generated strand-specific maps of R-loops, at base-pair resolution, using cross-linking and analysis of cDNA (CRAC). We used catalytically dead RNase H1-D193N as a bait to “fish” for R-loops. We tagged wild-type (WT) and RNase H1-D193N for immuno-purification, and the gene was put under the control of the Gal promoter for over-expression of RNase H1 in galactose. We also used the catalytic subunit of RNA polymerase II (RNAP II) as a bait to analyze the nascent transcriptome at mRNA genes and non-coding RNAs (ncRNAs). In the absence of Rrp6, a co-factor of the RNA exosome, cryptic and ncRNAs accumulate, which can form R-loops when RNase H1 is inactive. We are in the process of identified the RNA/DNA hybrids that are presents in these cells.
Separation of functions of RNase H2
The functions of RNase H2 share the same catalytic cleavage mechanisms as those of RNase H1, but have slightly different recognition of substrates. Humans with mutations in RNase H2 exhibit a severe interferonopathy, and each of its activities could contribute to the symptoms of the disorder. Mouse and human RNases H2 recognize and cleave the RNA of RNA/DNA hybrids. However, RNase H2 has a strong preference for hydrolysis at the rNMP–dNMP junction, as seen when single rNMPs are incorporated during DNA replication and when RNAs serve as primers of DNA synthesis and repair. We previously constructed a mutated form of RNase H2 almost completely defective for junction cleavage activity but retaining significant RNA/DNA hydrolysis. Mice bearing the mutated form die at the same early embryonic stage as the result of a complete loss of function of RNase H2. We constructed a mutated form of RNase H2 that retains high levels of junction activity and only a few percent activity of RNA/DNA hydrolysis. Mice carrying this mutation are indistinguishable from WT mice, at least as far as we have tested. The phenotypes of these two types of mice support the hypothesis that rNMPs incorporated in DNA, if retained, are extremely deleterious to cells and therefore organisms.
A mouse model based on a mutated RNase H2 found in some Aicardi-Goutières syndrome (AGS) patients with RNASEH2A Gly37-to-Ser replacement
No patient has been described with complete loss of either or both functions of RNase H2. One of the most defective RNases H2 described in AGS patients has a Gly amino acid replaced with Ser in the catalytic subunit. Our original RNase H2A–G37S knock-in mouse strain produced only perinatal lethal pups. Homozygous G37S embryos were about 70% the size of WT embryos from as early as E10 and remained similarly small until birth. After some time, we obtained a few viable mice, which we then bred, yielding non-Mendelian frequencies of homozygous offspring. The G37S knock-in mice were generated in a C57BL/6N (a common inbred mouse strain) background. We posited that crossing the original strain to C57BL/6J WT mice would reveal modifier genes to account for the viable births. We generated a congenic line with the G37S change in a C57BL/6J background and found no viable pups, supporting the modifier-gene hypothesis. A white ventral spotting of various sizes and shapes was present in more than 99% of the viable G37S mice. The spotting indicates a defect in the neural crest, most likely attributable to the production of insufficient numbers of melanocytes and/or insufficient energy to transport them to the belly. The white spotting and small size are seen in mice with haploinsufficiency in any one of several genes encoding ribosomal proteins that were generated to mimic the human disorder Diamond-Black-Fan anemia (DBA). Most DBA mice do not exhibit anemia, as is the case for our G37S mice. Our RNA-Seq data from G37S homozygous adult livers does indicate a defect in energy, as evidenced by defects in the mTOR pathway, the major pathway involved in energy metabolism. The level of L-leucine can have a positive influence on the mTOR pathway. Therefore, we provided our mice with drinking water containing L-leucine. We found no change in body weight (small size) but did see decreases in white-spotting size and several instances of no white spot. Defects in splicing often account for phenotypes. We will examine mouse embryonic fibroblasts for splice variations. We will also perform RNA-Seq analysis on the G37S congenic strain.
Mouse RNase H1: separation of function mutations
We found that transcription of the mouse Rnaseh1 gene produces a single transcript that is translated to make a protein targeted to mitochondria and a second protein that is in nuclei. Upon entering mitochondria, the N-terminal sequence of the protein is removed, making both the mitochondrial and the nuclear forms of RNase H1 identical. Mice with loss of the Rnaseh1 gene are embryonic lethal, because they fail to make mitochondrial DNA. We manipulated the Rnaseh1 gene to limit its expression to producing only the mitochondrial form. Mice that are homozygous for the mutation are viable and fertile. RNA-Seq data appear to mirror that of WT mice.
Publications
- Two RNase H2 mutants with differential rNMP processing activity reveal a threshold of ribonucleotide tolerance in DNA for embryonic development. Cell Rep 2018 25:1135–1145
- High density of unrepaired genomic ribonucleotides leads to Topoisomerase 1-mediated severe growth defects in absence of ribonucleotide reductase. Nucleic Acids Res 2020 48:4274–4297
- RNA abasic sites in yeast and human cells. Proc Natl Acad Sci USA 2020 117:20689–20695
- A common transcriptional mechanism involving R-loop and RNA abasic site regulates an enhancer RNA of APOE. Nucleic Acids Res 2022 50(21):12497–12514
Collaborators
- Charles Bou-Nader, PhD, Emory University, Atlanta, GA
- Frederic Chedin, PhD, University of California Davis, Davis, CA
- Louis Dye, BS, Microscopy and Imaging Core, NICHD, Bethesda, MD
- Aziz El Hage, PhD, University of Edinburgh, Edinburgh, United Kingdom
- James Iben, PhD, Molecular Genomics Core, NICHD, Bethesda, MD
- David Tollervey, PhD, University of Edinburgh, Edinburgh, United Kingdom
- Jinwei Zhang, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
Contact
For more information, email crouchr@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/crouch.