You are here: Home > Program on Pediatric Imaging and Tissue Sciences
Quantitative Biophotonics for Tissue Characterization and Function

- Amir H. Gandjbakhche, PhD, Head, Section on Analytical and Functional Biophotonics
- Victor Chernomordik, PhD, Staff Scientist
- Jason Riley, PhD, Research Fellow
- Moinuddin Hassan, PhD, Research Fellow
- Jana Kainerstorfer, PhD, Postdoctoral Fellow
- Laleh Najafizadeh, PhD, Postdoctoral Fellow
- Yasaman Ardeshirpour, PhD, Postdoctoral Fellow
- Franck Amyot, PhD, Guest Researcher
- Alexander Sviridov, PhD, Guest Researcher
- Atieh Bakhtiar, Volunteer
- Fatima A. Chowdhry, MD, Volunteer
- Bahar Dasgeb, MD, Volunteer
- Nader Shahni Karamzadeh, Volunteer
- Rizwan Jagani, Summer Student
- Michael Kellman, Summer Student
- Krishna Nagineni, Summer Student
- Sweetha Mannem, Summer Student
- Rafa Rahman, Summer student
- Rustin Tashayyod, Summer Student
- Sina Zarghan, Summer Student
Our objectives are to devise quantitative biophotonics methodologies and associated instrumentations in order to study biological phenomena at different length scales—from nanoscopy to microscopy—and diffuse biophotonics. We take advantage of our expertise in stochastic modeling to study complex biological phenomena whose properties are characterized by elements of randomness in both time and space, such as light-tissue interactions. We explore various properties of light-matter interactions as sources of optical contrasts, such as polarization properties, endogenous or exogenous fluorescent labels, absorption (e.g., hemoglobin or chromophore concentration), and/or scattering. We have used these contrast mechanisms for tomographic and spectroscopic methods to develop benchtop instrumentation for preclinical and clinical uses. We are identifying physiological sites where optical techniques might be clinically practical and offer new diagnostic knowledge and/or less morbidity than existing diagnostic methods.
Quantitative characterization of tissue
Polarization imaging is a promising tool to visualize hidden structures below the tissue surface. Analysis of such structures as, for example, the collagen network can be used to assess the possible transition from normal tissue function to diseased tissue. We developed sophisticated techniques of data analysis, based on the Pearson correlation procedure, that permit enhancement of image quality and that reveal regions of high statistical similarities within the noisy images, enabling characterization of subsurface structural features of biological tissues. To realize the potential of the method, we designed a user-friendly polarization imaging system that simultaneously images cross- and co-polarized light. Preliminary experiments demonstrated that the designed polarization adapter in combination with data analysis algorithms can provide quantitative information on collagen structure. We have incorporated this adapter into a conventional colposcope and started a clinical protocol to study tissue structures in the cervix. We collected data on 10 healthy volunteers at luteal and follicular phases of the cycle and are analyzing them to optimize the system and update the clinical protocol.
Preclinical studies on fluorescence imaging of HER2–positive breast cancer
Click image to enlarge.
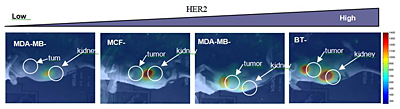
Figure 1. Accumulation of ZHER2- in breast carcinoma xenografts
Cell lines with different HER2 expression levels
The use of fluorescently labeled cell surface markers has led to a rapid advancement in specific molecular diagnosis of disease by histopathology and has already contributed to our understanding of the molecular origins of disease progression. Fluorophores, conjugated to such engineered molecules can target given cell receptors (e.g., human epidermal growth factor receptor type 2 [HER2]) and can be used for in vivo imaging. Such fluorescent probes preferentially concentrate at the sites of interest (e.g., in the tumor) after injection, which potentially permits quantification of HER2 expression in vivo. Probe accumulation parallels the delivery of some HER2–specific drugs, e.g., Trastuzumab, to the diseased site and opens a new era of "treat and image." It has been shown that overexpression of HER2, a well-known biomarker, plays an important role in aggressive tumor behavior and is associated with poor clinical outcome, confirming the importance of early characterization of HER2 receptors in the tumor. Current techniques for estimating HER2 receptor expression use ex vivo assays that require tissue biopsies. Furthermore, although it would be important, especially at the early stages, to continuously monitor of the response of tumor receptors to therapy to quantify the drug binding process—critical to timely evaluation of the drug efficacy—frequently lacking in cancer treatment assessment (for example, using monoclonal antibodies [MAB]). We have suggested a novel noninvasive method to characterize HER2 expression in vivo, using optical imaging and HER2–specific probes that could be used simultaneously with HER2–targeted therapy.
Further development of our project led us to assess HER2 overexpression in breast carcinoma xenografts in vivo (mouse model), using near-infrared fluorescence imaging. Novel specific probes based on small Affibody molecules in combination with in-house instrumentation allowed us to characterize tumors with varying levels of HER2 overexpression in the cancer cells in vivo (Figure 1). We tested two fluorescent dyes (AlexaFluor 750 and DyLight) with different binding/washout properties and showed that HER2 overexpression can be estimated from analysis of a series of the tumor fluorescence images, if the probe pharmacokinetics is taken into account by a ligand-receptor kinetic model. Experimental data for xenografts of a HER2–positive (3+) tumor BT474 indicate that the normalized initial rate of accumulation (NRA) of the contrast agent (probe) in the tumor depends linearly on the HER2 expression, as measured ex vivo by an ELISA assay for the same tumor. In a mouse model, we were able to monitor changes in the tumor resulting from treatment with a known anti-cancer drug 17-DMAG. As early reporters of down-regulation of receptors in response to molecular target therapy, the probes show promise for in vivo monitoring for HER2–based therapy.
Fluorescence lifetime imaging
Lifetime is another characteristic of fluorescence that can be estimated from time-resolved imaging, performed with our time-resolved instrumentation. With a proper dye it can potentially provide information about fluorophore environment (pH, temperature, tissue oxygen content, nutrient supply, and bioenergetic status) that can be of real diagnostic value. In collaboration with Samuel Achilefu and Gary Griffiths, we recently developed a novel pH–sensitive dye and tested it in phantoms. We showed that one of the developed dyes is sensitive to pH in the clinically important range of pH 5.5–7.0 (normal tissue pH range is 7.0–7.5, but in many solid tumors it can be as low as 5.6). We are also investigating changes in the fluorescence lifetime for HER2–specific probes in the tumor in vivo (mouse model) in order to assess the properties of cancer cells. Such information may be important to optimize treatment.
We use our in-house small-animal imaging system to develop more accurate methods of 3-D localization of the fluorophores inside turbid media. Application of the general analytical model, developed on the basis of a random walk theory in collaboration with George Weiss and Sinisa Pajevic, permitted accurate reconstruction of intrinsic fluorescence lifetime and estimation of the depth of an embedded inclusion in tissue-like phantoms.
Multispectral imaging
The oncology community is testing several novel targeted approaches for the treatment of a variety of cancers. With regard to monitoring vasculature, it is desirable to develop and assess noninvasive and quantitative techniques that can not only monitor structural changes but can also assess the functional characteristics or the metabolic status of the tumor. We are testing three potential noninvasive imaging techniques to monitor patients undergoing experimental therapy: infrared thermal imaging (thermography), laser Doppler imaging (LDI), and multi-spectral imaging. We are testing the imaging techniques on subjects with Kaposi's sarcoma (KS), a highly vascular tumor that occurs frequently among patients infected with HIV. Cutaneous KS lesions are easily accessible for noninvasive techniques to image tumor vasculature and thus represent a tumor model in which to assess certain parameters of angiogenesis. We are pursuing KS studies in ongoing clinical trials under four different NCI protocols. We have shown that our multi-modality techniques can non-invasively monitor the functional properties of the tumor and surrounding tissues and have the potential to predict treatment outcomes. We recently obtained angiogenic markers (blood volume and oxygenation) in real time. Using novel data analysis tools based on Principal Component Analysis, we showed that a KS lesion can be imaged and the functional state of the tumor assessed in real time. In combination with optical coherence tomography (OCT), we recently obtained quantitative measures of blood volume and oxygenation in real time. We also found that quantitative results can be obtained if the underlying skin structures are taken into account. In order to obtain those structures, we developed a spectral domain OCT system that provides 3D images of the skin.
Structural and functional brain imaging
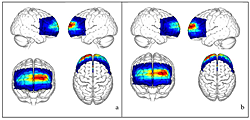
Click image to enlarge.
Figure 2. HbR (a) and HbO (b) concentration maps on the surface cortex of the MNI atlas
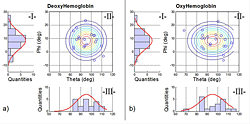
Click image to enlarge.
Figure 3. Normal angular distribution for (a) HbR and (b) HbO for the 20 subjects
The 2-D angular distribution (II-a and II-b) is given by the normal distribution of elevation (red curve in I-a and I-b) and the normal distribution of the azimuth (red curve III-a and III-b). The lines in Figure II-a and II-b are the iso-probabilities lines calculated; the blue circles represent the subject activation coordinate.
The frontal lobes are particularly vulnerable to traumatic brain injury. However, in mild cases, lack of sensitive bedside tests for executive function can make it hard to asses frontal lobe involvement, which may not even be suspected in survivors who are otherwise neurologically intact. Thus, casualties with occult frontal impairment and deficits in judgment may even be discharged inappropriately. There is therefore a need for reliable tests of frontal lobe function that can be administered quickly by individuals without neuropsychological training. We seat subjects on an adjustable chair, in front of a computer that displays a stimulus and apply a functional near-infrared spectroscopy (fNIRS) detector/emitter array to the appropriate forehead position; the array is held in place by on an adjustable band. Each fNIRS trial starts with an instruction to judge "complexity" or "font" in the stimulus. In complexity trials, participants have to signal a judgment (few events vs. many) about the complexity of the activity, using a response pad. In "font" trials, we ask subjects to decide whether the trial instruction text was presented in the same font as the activity stimulus. We recruited 20 adults for this study. Each participant underwent a previous MRI scan, which was registered with the MNI (Montreal Neurological Institute) atlas. We compared fNIRS data from each channel acquired from font and complexity tasks. We also looked for the association between stimulus complexity and the magnitude of blood–flow increase in the frontopolar cortex (Figure 2). For each experiment, we co-registered all channels with the MRI anatomical patient image using a stereotactic camera. For the group study, we found activation at the coordinates x = -7, y = 85, z = 13 in the MNI atlas, which is consistent with results from functional magnetic resonance imaging (fMRI) studies. From the subject measurements, we constructed a normative database for a simple cognitive test that can be useful in evaluating cognitive disability such as mild TBI (Figure 3). We are now investigating the correlation between the behavioral data and the delay in hemodynamic response. Our last aim is to study the parametric design of our paradigm and determine whether frontopolar cortex activity correlates with complexity based on the normative rating for each daily-life event. In summary, we used a simple task to produce increased anterior frontal lobe blood flow and, by using MRI co-registration, group analysis, and an angular mapping technique, showed that the scalp activity detected with a simple NIRS apparatus corresponded to the blood flow change detected with fMRI during the same task.
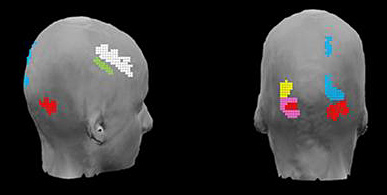
Figure 4. Captured connectivity maps using EEG
EEG in response to an n-back working memory task. Side and back view.
We have also been involved in studying the dynamics of brain interactions on two different scales by combing electroencephalography (EEG) and diffuse optical imaging (DOI) techniques. Simultaneous recordings of DOI and EEG signals would allow us to identify connectivity among areas of the brain that are involved in executing a certain task at two temporal scales and also to study neurovascular coupling by correlating the results from neural recording by EEG with the hemodynamic response captured by DOI. Additionally, we have been working on developing new analysis techniques to identify brain functional connectivity (both task-based and in the resting state) from EEG and DOI data. The existing techniques such as correlation and data-driven decompositions, which are employed to assess the functional connectivity, generally assume temporally stationary brain signals over the duration of the recording. Such an assumption may result in dismissing dynamic changes. We have been working on applying time-frequency analysis and clustering techniques to capture the dynamic behavior of connectivity. Initial results are shown in Figure 4, which illustrates EEG–captured connectivity in response to an n-back task.
Additional Funding
- Bench to Bedside Award
- Center for Neuroscience and Regenerative Medicine
- NIBIB
Publications
- Kainerstorfer JM, Riley JD, Ehler M, Najafizadeh L, Amyot F, Hassan M, Pursley R, Demos SG, Chernomordik V, Pircher M, Smith PD, Hitzenberger CK, Gandjbakhche AH. Quantitative principal component model for skin chromophore mapping using multi spectral images and spatial priors. Biomedical Optics Express 2011;1040-1058.
- Berezin MY, Guo K, Akers W, Northdurft RE, Culver JP, Teng B, Vasalatiy O, Barbacow K, Gandjbakhche A, Griffiths GL, Achilefu S. Near-infrared fluorescence lifetime pH-sensitive probes. Biophys J 2011;14:2063-2072.
- Medvedev AV, Kainerstorfer JM, Borisov SV, Gandjbakhche AH, Vanmeter J. "Seeing" electroencephalogram through the skull: imaging prefrontal cortex with fast optical signal. J Biomed Opt 2010;061702.
- Hassan M, Chernomordik V, Zielinski R, Ardeshirpour Y, Capala J, Gandjbakhche A. In vivo method to monitor changes in HER2 expression using near-infrared fluorescence imaging. Mol Imaging 2011;in press.
- Combs CA, Smirnov A, Chess D, McGavern DB, Schroeder JL, Riley J, Kang SS, Lugar-Hammer M, Gandjbakhche A, Knutson JR, Balaban RS. Optimizing multiphoton fluorescence microscopy light collection from living tissue by noncontact total emission detection. J Microsc 2011;153-161.
Collaborators
- Samuel Achilefu, PhD, Washington University School of Medicine, St. Louis, MO
- Robert Balaban, PhD, Laboratory of Cardiac Energetics, NHLBI, Bethesda, MD
- Claude Boccara, PhD, École Supérieure de Physique et de Chimie Industrielles, Paris, France
- Kevin Camphausen, PhD, Radiation Oncology Branch, NCI, Bethesda, MD
- Jacek Capala, PhD, Radiation Oncology Branch, NCI, Bethesda, MD
- Christian Combs, PhD, Light Microscopy Facility, NHLBI, Bethesda, MD
- Stavros Demos, PhD, Lawrence Livermore National Laboratory, Livermore, CA
- Israel Gannot, PhD, Tel Aviv University, Tel Aviv, Israel, and The George Washington University, Washington, DC
- Gary Griffiths, PhD, Imaging Probe Development Center, NIH, Rockville, MD
- Ilko Ilev, PhD, Office of Science and Engineering Laboratories, FDA, Bethesda, MD
- Jay Knutson, PhD, Laboratory of Molecular Biophysics, NHLBI, Bethesda, MD
- Ralph Nossal, PhD, Program in Physical Biology, NICHD, Bethesda, MD
- Sinisa Pajevic, PhD, Division of Computational Bioscience, CIT, NIH, Bethesda, MD
- Dan Sackett, PhD, Program in Physical Biology, NICHD, Bethesda, MD
- Alexsandr Smirnov, PhD, Light Microscopy Facility, NHLBI, Bethesda, MD
- Paul Smith, PhD, Division of Bioengineering and Physical Science, ORS, Bethesda, MD
- Eric Wassermann, MD, Cognitive Neuroscience Section, NINDS, Bethesda, MD
- George Weiss, PhD, Division of Computational Bioscience, CIT, NIH, Bethesda, MD
- Robert Yarchoan, MD, HIV and AIDS Malignancy Branch, NCI, Bethesda, MD
- Rafal Zielinski, PhD, Radiation Oncology Branch, NCI, Bethesda, MD
Contact
For more information, email amir@helix.nih.gov.

