You are here: Home > Program on Pediatric Imaging and Tissue Sciences
Tissue Biophysics and Biomimetics

- Peter J. Basser, PhD, Head, Section on Tissue Biophysics and Biomimetics
- Ferenc Horkay, PhD, Staff Scientist
- Carlo Pierpaoli, MD, PhD, Staff Scientist
- Okan Irfanoglu, PhD, Henry M. Jackson Foundation Contractor
- Michal Komlosh, PhD, Henry M. Jackson Foundation Contractor
- Evren Özarslan, PhD, Henry M. Jackson Foundation Contractor
- Iren Horkayne-Szakaly, MD, Volunteer
- Candida Silva, PhD, Volunteer
- Amritha Nayak, Technical Intramural Research Training Awardee
- Lindsay Walker, MS, Guest Researcher
- Pooja Modi, BS, Postbaccalaureate Intramural Research Training Award Fellow
We attempt to understand fundamental relationships between function and structure in living tissues, "engineered" tissue constructs, and tissue analogues. Specifically, we are interested in how microstructure, hierarchical organization, composition, and material properties of tissue affect its biological function and dysfunction. We investigate biological and physical model systems at different time and length scales, making physical measurements in tandem with the development of mathematical and computational models. Primarily, we use water molecules to probe both equilibrium and dynamic interactions among tissue constituents over a wide range of time and length scales. To determine the equilibrium osmo-mechanical properties of well defined model systems, we vary water content or ionic composition. To probe tissue structure and dynamics, we employ atomic force microscopy (AFM), small-angle X-ray scattering (SAXS), small-angle neutron scattering (SANS), static light scattering (SLS), dynamic light scattering (DLS), and nuclear magnetic resonance (NMR) relaxometry. We develop and use mathematical models to help us understand how observed changes in tissue microstructure and physical properties (e.g., mass, charge, and momentum) affect essential transport processes. The most direct noninvasive in vivo method for characterizing these transport processes in tissues is magnetic resonance imaging (MRI), which we use to follow microstructural changes in development, degeneration, aging, and trauma. A goal of our basic tissue sciences research is to translate our quantitative methodologies and the understanding we glean from them from "blackboard to bench to bedside."
Virtual in vivo histology
We aim to develop novel next-generation in vivo imaging methods to help us gain a better understanding of brain function and organization and to improve the diagnosis of neurological and developmental disorders. To these ends, the most mature technology that we have invented and developed is Diffusion Tensor MRI (DTI). With it, we measure a diffusion tensor of water, D, voxel-by-voxel within an imaging volume. Information derived from D includes the local nerve fiber–tract orientation, the mean-squared distance that water molecules diffuse in each direction, the orientationally averaged mean diffusivity, and other intrinsic scalar (invariant) quantities (i.e., those that are independent of the laboratory coordinate frame used to make these measurements). These scalar parameters derived from D behave like quantitative histological "stains." However, they are obtained from endogenous tissue water and are "developed" without exogenous contrast agents or dyes. The bulk or orientationally averaged diffusivity is the most successful DTI parameter proposed to date for identifying ischemic regions in the brain during acute stroke. Measures of diffusion anisotropy (e.g., the "fractional anisotropy" or FA) are widely used to follow changes in normally and abnormally developing white matter, including dysmyelination and demyelination. Our group also pioneered the use of fiber direction–encoded color (DEC) maps (derived from D) to reveal nerve fiber orientation, which can be used to identify the main association, projection, and commissural white matter pathways in the brain. To assess anatomical connectivity among various functional brain regions, we also developed DTI "streamline" Tractography to track nerve fiber trajectories by continuously following the direction along which the apparent diffusivity is maximal.
More recently, we invented and have been further developing several advanced in vivo MR methods to measure fine-microstructural features of nerve fascicles, which previously could be measured only by optical microscopy using laborious ex vivo histological methods. We are developing efficient methods for performing "k and q-space MRI" in the living brain. This approach can detect subtle microstructural and architectural features in both gray and white matter at a finer spatial resolution than DTI. We can also use this approach to characterize features of "anomalous" or fractal diffusion that we recently observed in the neuropil. We also previously developed a diffusion MRI method to measure the axon-diameter distribution (ADD) within large white-matter fascicles, dubbing the method AxCaliber MRI. After careful validation studies, we recently reported the first in vivo measurement of axon diameter distributions within the corpus callosum in the rodent brain. The ADD is important neurophysiologically and developmentally given that axon diameter determines nerve conduction velocity and therefore the rate of information transfer along nerve pathways. We developed mathematical models in tandem with these experiments to help discover important organizing principles that underlie observed axon diameter distributions in different fascicles and features of information transfer between various brain regions. Novel multiple pulsed-field gradient (PFG) methods that we developed are providing us with independent measurements of mean axon diameter, making the performance of these sequences feasible on conventional clinical MRI scanners and thus rapidly translatable.
While gray matter appears featureless in DTI, its microarchitecture is rich and varied, not only along the cortical surface but also within different cortical layers. We have been developing several noninvasive, in vivo methods to "drill down" into the image voxel to measure unique features of cortical gray matter microstructure that are currently invisible in conventional MRI. One goal is to "parcellate" or segment the cerebral cortex in vivo into its distinct "Brodmann" areas. To this end, we are developing advanced MRI sequences to probe correlations among microscopic displacements of water molecules in the neuropil as well as sophisticated mathematical models to infer distinguishing microstructural and morphological features of gray matter. We are continuing to develop these methods, which will permit us to follow normal and abnormal development of the cerebral cortex noninvasively and in vivo.
Quantitative pediatric MRI
MRI is considered safer for non-invasively scanning pediatric subjects than x-ray based methods such as Computed Tomography (CT). Traditionally, clinical MRI has relied on the acquisition of so called "weighted images," whose contrast is affected by many factors, some intrinsic to the tissue, and some dependent on the details of the experiment and experimental design. The diagnostic utility of conventional MRI in many neurological disorders is unquestionable. However, the scope of conventional MRI applications is limited to revealing either gross morphological or focal abnormalities resulting in regional differences in signal intensities within a given tissue. To detect pathology, conventional MRI relies on differences in contrast between areas that are supposedly affected and supposedly normal. Thus, it is intrinsically insensitive to subtle global changes that may affect the entire tissue. Clinical MRI also lacks biological specificity. Although quantification per se does not assure such specificity, it is necessary for developing imaging "biomarkers." The MRI assessment of normal brain development and developmental disorders is benefiting greatly from "quantitative" clinical MRI techniques, in which one obtains maps of meaningful physical quantities or chemical variables that can be measured in physical units and compared among and between tissue regions, in longitudinal and cross-sectional studies. Quantitative MRI also increases sensitivity, providing a basis for monitoring subtle changes that occur during the progression or remission of disease by comparing measurements in a single subject with normative values acquired in a healthy population. Finally, quantitative MRI methods should enhance our ability to perform comparative effectiveness research for a variety of diagnosis and therapy assessments.
Our group is carrying out several clinical studies that utilize novel quantitative MRI acquisition and analysis methods, whose aim is to improve accuracy and reproducibility in diagnosis and to detect and follow normal and abnormal development. The studies include:
- The NIH Study of Normal Brain Development, jointly sponsored by four NIH Institutes (NICHD, NIMH, NINDS, and NIDA) is a multicenter study to advance our understanding of brain development in typical healthy children and in adolescents. Structural MR images and the results of standardized neuropsychological tests from this population are now available to researchers. Our role in this interdisciplinary project has been to serve as a DTI Data-Processing Center (DPC). We have now processed all admissible DTI data and have released them to a database accessible to all interested investigators. We continue to analyze the data, developing age-specific DTI atlases of normal brain development. We also released publicly the software we developed for this project and related documentation, which can be downloaded from http://www.tortoisedti.org. In particular, we are using our advanced DTI–processing pipeline to produce high-quality normative data from this project and to make them publicly available through the National Database for Autism Research (NDAR; https://nda.nih.gov).
- In collaboration with Susan Swedo (NIMH), we have studied autistic subjects using DTI and quantitative relaxometry methods. Several MRI studies have reported abnormal features in the autistic brain, but no clear MRI "biomarker" of autism exists. The aim of this study is to use robust quantitative metrics to identify potential anatomical abnormalities in the autistic brain and to find candidate imaging biomarkers for the disorder.
- In collaboration with Kathy Warren (NCI), we acquired quantitative MRI data in children with pontine gliomas to identify MRI prognostic factors. With John Park (NINDS), we are scanning subjects with supratentorial gliomas to distinguish between recurrence and radiation necrosis.
- In collaboration with Filippo Arrigoni (Institute "Eugenio Medea," (http://www.emedea.it/english_medea/), we are using multimodality MR imaging (DTI, fMRI, and quantitative relaxometry) to evaluate cerebral reorganization caused by various rehabilitation protocols in children with cerebral palsy and traumatic brain injury (TBI).
- In addition to these clinical studies, we are working under the auspices of the Center for Neuroscience and Regenerative Medicine (CNRM, http://www.usuhs.mil/cnrm/) to develop enhanced software tools for analysis of diffusion MRI in TBI and post-traumatic stress disorder (PTSD).
- In collaboration with Sharon Juliano (Uniformed Services University of the Health Sciences), we submitted a grant proposal to the Congressionally Directed Medical Research Program (CDMRP) to investigate TBI in the ferret using advanced MRI and histopathology techniques. The proposal was awarded funding, and we will start preliminary experiments in early 2013.
Our involvement in TBI research is expanding. DTI provides essential information to diagnose TBI and could be developed into an important tool for the assessment of potential structural damage in PTSD. For clinical applications, however, we need reliable imaging protocols. Part of our work is to develop a robust DTI data–processing pipeline in order to improve the accuracy and reproducibility of DTI findings for CNRM investigators and potentially for the larger clinical and scientific community involved in TBI research. To this end, we are adding new modules to our existing state-of-the-art DTI data–processing pipeline as well as tools to permit calibration of DTI experiments using a new polymer-based diffusion MRI phantom that we developed and are disseminating to clinical sites.
Click image to enlarge.
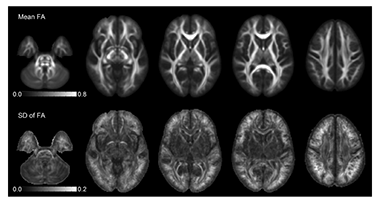
Figure 1. High-quality DTI "Atlases" can now be obtained with relatively small subject populations.
Maps of the mean and standard deviation of Fractional Anisotropy (FA) obtained from a population of normal pediatric subjects (N=40) after diffeomorphic tensor registration. This improved registration scheme reduces voxelwise variability as compared with conventional affine registration (not shown), particularly in the major white matter tracts and in the cerebellum. Higher variability can be observed in peripheral regions, particularly at the top of the brain.
Looking ahead, to permit analysis of novel MRI data sets such as the ones described above, as well as new clinical and biological applications of quantitative MRI, we need to create a new mathematical, statistical, and image-sciences infrastructure. To date, we have developed algorithms that generate a continuous, smooth approximation of the discrete, noisy, measured DTI field data so as to reduce noise and permit us to follow fiber tracts more reliably. We proposed a new Gaussian distribution for tensor-valued random variables that we used in designing optimal DTI experiments. In tandem, we developed nonparametric empirical (e.g., Bootstrap) methods for determining the statistical distribution of DTI–derived quantities in order to study, for example, the inherent variability and reliability of white matter fiber tract trajectories. These parametric and nonparametric statistical methods will enable us to apply powerful hypothesis tests to assess statistical significance in a wide range of important biological and clinical questions that are currently being addressed using ad hoc methods. We are also developing novel methods to generate group-average data or atlases from various subject populations. We are also developing methods for studying the reproducibility and reliability of tractography findings (Figure 1). However, much work remains to be done in order to permit statistically significant inferences from clinical DTI data, particularly those obtained in longitudinal and multicenter studies.
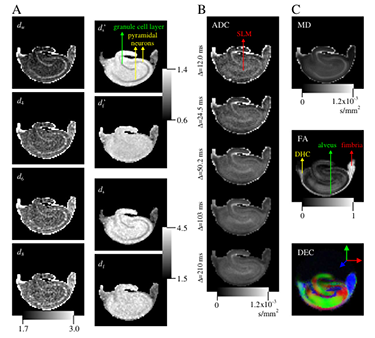
Figure 2. Novel fractal diffusion model provides new "stains" to visualize tissue microstructure.
A: Images of the exponents characterizing the fractal diffusion process in the rat hippocampus. B: The apparent diffusion coefficient (ADC) maps for five diffusion times computed from the low-q (DTI) regime. C: Traditional DTI–derived mean diffusivity (MD), fractional anisotropy (FA), and direction-encoded color (DEC) maps computed for the same slice. White-matter structures of dorsal hippocampal commissure (DHC), alveus, and fimbria are identifiable on the FA map, whereas the regions of granule cell layer and pyramidal neurons are shown on the d maps. The SLM label on the ADC map stands for stratum lacunosum-moleculare.
Biopolymer physics: water-ion-biopolymer interactions
Click image to enlarge.
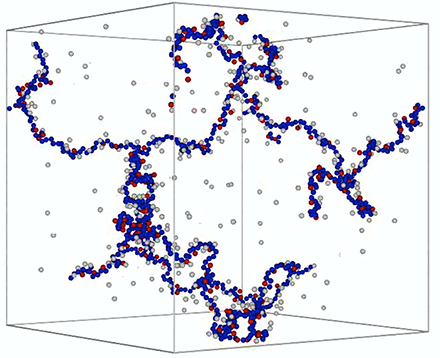
Figure 3. Simulation of ion-water-biopolymer interactions reveal different behaviors of mono- and divalent counterions.
Snapshot of a simulated polyelectrolyte gel in an aqueous solution with mono- and divalent counterions. The polymer backbone is shown in blue, monovalent counterions in gray, and divalent counterions in red. The figure illustrates that divalent counterions condense onto the polymer, while a significant fraction of the monovalent counterions remains free in solution.
Our objective is to better understand water-ion-biopolymer interactions in biological systems, such as polynucleic acids, proteins, and membranes—interactions that often play a dominant role in determining system function and behavior. The research promises to generate new insights into the interactions' physical and chemical bases; a fundamental understanding of the nature of non-covalent molecular interactions operating between ions and biopolymers has already helped us optimize the stability of novel nano-structures (e.g., DNA–based "nano-medicine"), create scaffolds for tissue regeneration, and enhance the performance (e.g., selectivity) of glucose sensors.
We developed an experimental approach to study the structure (morphology) and thermodynamic properties of biopolymer assemblies as a function of the length scale (spatial resolution) by combining macroscopic osmotic swelling pressure measurements and small-angle scattering measurements. Swelling pressure measurements probe the system at large length scales, providing information on its overall thermodynamic response, while SANS and SAXS methods allow us to investigate native biopolymer molecules. Combining measurements of changes in physical properties, such as molecular conformation and osmotic pressure, with changes in environmental conditions, such as ion concentration, ion valance, pH, and temperature, helps us discover the length scales that govern key macroscopic thermodynamic properties. This information cannot be obtained by other techniques.
Divalent cations, particularly Ca2+ ions, are ubiquitous in the biological milieu; yet existing theories of polyelectolytes do not adequately explain or account for their effect on and interactions with charged macromolecules. In pilot studies, we used our new non-destructive procedure to investigate the effect of Ca2+ ions on hyaluronic acid (HA) and DNA gels. The gel systems also mimic certain essential features of the extracellular matrix (ECM) and have potential as scaffolds for tissue-regeneration applications.
We investigated the binding mechanism in glucose sensors made from smart zwitterionic hydrogels containing boronic acid moieties. Based on systematic SANS and osmotic pressure measurements, we provided a thermodynamic explanation for the enhanced selectivity of the gels for glucose relative to fructose. This class of material has great potential for the development of implantable continuous glucose sensors for use in Type I and Type II diabetes.
We also developed a method to control the size, compactness, and stability of DNA nano-particles by mediating the interaction between ions and DNA. We quantified the effects of salt, pH, and temperature on the nano-particles' stability and biological activity. Such polyplexes are pathogen-like particles, 70–300 nm in size, and with shapes resembling spherical viruses that evolved naturally to deliver nucleic acids to cells. The nano-particles contain the pDNA in an interior surrounded by a synthetic polymer bearing surface sugar residues, which are recognized by the "M" cells and dendritic cells as belonging to pathogens. We determine and tailor the physical and biological properties of the nano-particles, including their thermodynamic stability, knowledge that is essential in order to design and control properties of DNA–based nano-medicines. The biological activity of such nano-medicine depends on the competing requirements of sufficient stability to escape endosomal degradation after cellular uptake with retention of biological activity upon arrival in the nucleus, where the nano-particle must unravel and release the pDNA to achieve gene expression. By changing the sequence of the pDNA specific to the targeted disease, DNA–based vaccines may be promising as therapies in several disease indications, including infectious diseases, cancer, and allergies.
Functional properties of extracellular matrix
Click image to enlarge.
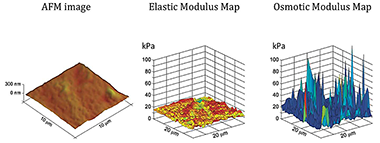
Figure 4. Microscale measurements of tissue material properties reveal significant heterogeneity.
AFM topographic image (left), elastic modulus (middle), and osmotic modulus (right) maps of a biomimetic tissue phantom (PVA hydrogel). Knowledge of the elastic and osmotic properties are particularly important to better understanding the basis of a tissue's functional properties, and how these may change in growth and repair, or change in development and disease. The modulus maps reveal fine-scale variations in the tissue's compressive resistance owing to changes in the structure and interactions among tissue constituents.
The biomechanical behavior of cartilage and other ECMs reflects both biochemical and microstructural changes occurring during development, disease, degeneration, and aging. To study the physical properties of cartilage (e.g., osmotic swelling properties and hydration), one requires an array of techniques that can probe a wide range of length and time scales. Our goal is to explain functional properties of cartilage and other ECMs in terms of their hierarchical structure, their key molecular components, and interactions among them. Understanding the physical and chemical mechanisms affecting cartilage swelling is essential to predicting critical functional properties—particularly its load-bearing and lubricating abilities—which are governed by osmotic and electrostatic forces that depend strongly on tissue hydration. Such understanding is also a prerequisite for the success of tissue engineering or of regenerative medicine strategies to grow, repair, and reintegrate cartilage. To gain a self-consistent physical picture of tissue structure/function relationships, we measure various physical/chemical properties of tissues and tissue analogues at different length- and time-scales using a variety of complementary experimental techniques, e.g., osmometry, SANS, SAXS, neutron spin-echo (NSE), SLS, DLS, AFM, and fluorescence correlation spectroscopy (FCS).
Controlled tissue hydration provides a direct means of determining the patency and load-bearing ability of cartilage. Previously, we used controlled hydration to measure physical/chemical properties of the collagen network and of proteoglycans (PG) independently within cartilage. We developed and built a novel tissue micro-osmometer to perform such experiments in a practical, rapid, and eventually automated manner. The instrument can measure minute amounts of water absorbed by small tissue samples (less than 1 microgram) as a function of the equilibrium activity (pressure) of the surrounding water vapor. A quartz crystal detects the water uptake of a specimen. The high sensitivity of the crystal's resonance frequency to small changes in the amount of adsorbed water allows us to measure the mass uptake of a tiny tissue specimen precisely.
We used osmotic pressure measurements to determine the contributions of individual components of cartilage ECM (e.g., aggrecan, hyaluronic acid [HA], and collagen) to the total tissue swelling pressure. Our measurements on aggrecan-HA systems revealed that the osmotic modulus of the aggrecan-HA complex is greater than that of random assemblies of aggrecan bottlebrushes, providing direct evidence that the aggrecan–HA complex improves load bearing in cartilage. We also demonstrated that aggrecan–HA assemblies exhibit microgel-like behavior and a remarkable insensitivity to changes in the ionic environment, particularly to the Ca+2 concentration. The latter result is consistent with the role of aggrecan as a calcium ion reservoir mediating calcium metabolism in cartilage and bone.
We also developed a novel method for mapping the local elastic and osmotic properties of cartilage using AFM together with the tissue micro-osmometer. Many impediments that previously hindered the use of AFM to probe inhomogeneous biological tissues were addressed by this new development. The technique utilizes the precise scanning capabilities of AFM to generate large volumes of compliance data, from which the relevant elastic properties are extracted. We mapped the osmotic modulus of bovine cartilage samples by combining tissue micro-osmometry with force-deformation measurements made by the AFM. Knowledge of the local osmotic properties of cartilage is particularly important given that the osmotic modulus defines the compressive resistance to external load. We found that water retention is stronger in the surface zones and deep zones of cartilage, where collagen fibers are ordered, than in the middle zone, where they are randomly arranged. We constructed the elastic and osmotic modulus maps for the various layers. The maps, which are a combination of the elastic and swelling properties, exhibit large, fine-scale spatial variation, reflecting the highly heterogeneous character of the tissue.
Additional Funding
- Award G189AM from the Henry Jackson Foundation supports the STBB's project in "AxCaliber MRI," which is under the joint auspices of the NIH, the DoD, the Center for Neuroscience and Regenerative Medicine (CNRM), and the Uniformed Services University of the Health Sciences (USUHS).
- Award 60855 from the Henry Jackson Foundation supports STBB's project in "Radial Diffusion-Weighted MRI Acquisitions for Diffusion Tensor Imaging of TBI," which is under the joint auspices of the NIH, DoD, CNRM, and USUHS.
- Award 305500-1.01-60855 from the Henry Jackson Foundation supports STBB's project in "Enhanced Software Tools for the Analysis of Diffusion MRI in TBI and PTSD," which is under the joint auspices of the NIH, DoD, CNRM, and USUHS.
- Award 306135-2.01-60855 from the Henry Jackson Foundation supports STBB's project "Clinical Double Pulsed-Field Gradient (dPFG) MRI for Mild TBI Assessment," which is under the joint auspices of the NIH, DoD, CNRM, and USUHS.
Publications
- Özarslan E, Shepherd TM, Koay CG, Blackband SJ, Basser PJ. Temporal scaling characteristics of diffusion as a new MRI contrast: Findings in rat hippocampus. NeuroImage 2012;60:1380–1393.
- Horkay F, Basser PJ, Hecht A-M, Geissler E. Hierarchical organization of cartilage proteoglycans. Macromol Symp 2011;306-307:11–17.
- Walker L, Curry M, Nayak A, Lange N, Pierpaoli C, Brain Development Cooperative Group. A framework for the analysis of phantom data in multicenter diffusion tensor imaging studies. Hum Brain Mapp 2012;E-pub ahead of print.
- Irfanoglu MO, Walker L, Sarlls J, Marenco S, Pierpaoli C. Effects of image distortions originating from susceptibility variations and concomitant fields on diffusion MRI tractography results. Neuroimage 2012;61(1):275-288.
- Özarslan E, Komlosh ME, Lizak MJ, Horkay F, Basser PJ. Double pulsed field gradient (double-PFG) MR imaging (MRI) as a means to measure the size of plant cells. Magn Reson Chem 2011;49:S79–S84.
Collaborators
- Filippo Arrigoni, MD, Fondazione IRCCS Eugenio Medea, Bosisio Parini, Italy
- Alan Barnett, PhD, Clinical Brain Disorders Branch, NIMH, Bethesda, MD
- Madison Berl, PhD, Children's National Medical Center, Washington, DC
- Preethi Chandran, PhD, Division of Bioengineering and Physical Science, NIBIB, Bethesda, MD
- Yoram Cohen, PhD, Tel Aviv University, Tel Aviv, Israel
- Emilios Dimitriadis, PhD, Division of Bioengineering and Physical Science, NIBIB, Bethesda, MD
- Raisa Freidlin, MS, Computational Bioscience and Engineering Laboratory, CIT, NIH, Bethesda, MD
- Erik Geissler, PhD, CNRS, Université Joseph Fourier de Grenoble, Grenoble, France
- Anne-Marie Hecht, PhD, CNRS, Université Joseph Fourier de Grenoble, Grenoble, France
- Sharon Juliano, PhD, Uniformed Services University of the Health Sciences, Bethesda, MD
- Stefano Marenco, PhD, Clinical Brain Disorders Branch, NIMH, Bethesda, MD
- Pedro Miranda, PhD, Universidade de Lisboa, Lisbon, Portugal
- Sinisa Pajevic, PhD, Mathematical and Statistical Computing Laboratory, CIT, NIH, Bethesda, MD
- John Park, MD, Surgical Neurology Branch, NINDS, Bethesda, MD
- Bradley Roth, PhD, Oakland University Department of Physics, MI
- Susan Swedo, MD, Pediatrics and Developmental Neuroscience Branch, NIMH, Bethesda, MD
- Kathy Warren, MD, Pediatric Oncology Branch, NCI, Bethesda, MD
- Brain Development Cooperative Group, Various
Contact
For more information, email pjbasser@helix.nih.gov or visit stbb.nichd.nih.gov.