You are here: Home > Section on Organelle Biology
Dynamics of Membrane Trafficking, Sorting, and Compartmentalization Within Eukaryotic Cells

- Jennifer Lippincott-Schwartz, PhD, Head, Section on Organelle Biology
- Dylan Burnette, PhD, Postdoctoral Fellow
- Jennifer Gillette, PhD, Postdoctoral Fellow
- Kasturi Mitra, PhD, Postdoctoral Fellow
- Carolyn Ott, PhD, Postdoctoral Fellow
- Angelika Rambold, PhD, Postdoctoral Fellow
- Richa Rikhy, PhD, Postdoctoral Fellow
- Prasanna Satpute, PhD, Postdoctoral Fellow
- Prabuddha Sengupta, PhD, Postdoctoral Fellow
- Tijana Talisman, Ph.D, Postdoctoral Fellow
- Christian Wunder, PhD, Postdoctoral Fellow
- Yu Chen, PhD, Visiting Fellow
We investigate the global principles underlying secretory membrane trafficking, sorting, and compartmentalization within eukaryotic cells. We use live-cell imaging of green fluorescent protein (GFP) fusion proteins in combination with photobleaching and photoactivation techniques to investigate the subcellular localization, mobility, transport routes, and turnover of a variety of proteins with important roles in the organization and regulation of membrane trafficking and compartmentalization. To test mechanistic hypotheses about protein and organelle dynamics, we use quantitative measurements of these protein characteristics in kinetic modeling and simulation experiments. Among the topics under investigation are 1) membrane partitioning and its role in protein sorting and transport in the Golgi apparatus; 2) biogenesis and dynamics of unconventional organelles; 3) mitochondrial morphology and its regulation of cell cycle progression; and 4) cytoskeletal and endomembrane cross-talk in polarized epithelial cells, hematopoietic niche cells, and the developing Drosophila syncytial blastoderm embryo. We also devoted considerable effort to developing new fluorescence microscopy techniques for imaging fluorescently tagged proteins at near-molecular resolution.
Development of green fluorescent protein technology
Super-resolution techniques such as photoactivated localization microscopy (PALM) permit the imaging of fluorescent protein chimeras, revealing the organization of genetically expressed proteins on the nanoscale with a density of molecules high enough to provide structural context. The PALM method involves serial photoactivation and subsequent bleaching of numerous sparse subsets of photoactivated fluorescent protein molecules. A statistical fit of the point-spread function of individual molecules' centers of fluorescent emission then localizes the molecules at near-molecular resolution. The aggregate position information from all subsets is then assembled into a super-resolution image that isolates individual fluorescent molecules at high molecular densities (up to 105 molecules/µm2). We previously demonstrated PALM imaging of intracellular structures (including lysosomes, Golgi apparatus, and mitochondria) in cryo-prepared thin sections as well as imaging of vinculin and actin in fixed cells with TIRF (total internal reflection fluorescence) excitation along with correlative PALM/transmission electron microscopy of a mitochondrial marker protein. We recently helped develop a new photoactivatable fluorescent protein called PA-mCherry that switches from dark to red upon UV illumination. Using PA-mCherry and PA-GFP, we demonstrated two-color photoactivation imaging. The two-color approach was also used for PALM imaging of two different proteins within cells. We also used PALM in a new technique for single particle tracking (sptPALM) in living cells. This allowed protein diffusion and immobilization to be characterized at the single-molecule level.
Membrane partitioning and its role in protein sorting and transport within the Golgi apparatus
The Golgi apparatus processes and filters newly synthesized protein and lipid moving through the secretory pathway, but how it transfers secretory cargo through its six to eight flattened cisternae has remained unclear. To gain insight into this question, we analyzed various classes of cargo molecules within the Golgi stack after they were fluorescently pulse-labeled, and we quantitatively visualized them as they transited from the Golgi. Our results revealed an exponential loss of cargo from the entire Golgi rather than the linear pattern predicted by the classical model of cisternal maturation. Moreover, when transmembrane cargo entered the Golgi apparatus, it differentially partitioned between two membrane environments—processing domains enriched in Golgi enzymes and export domains capable of budding transport intermediates. Based on the results of our experiments, we constructed and tested a new model of intra-Golgi trafficking in which cargo molecules continuously partition between processing and export domains, each defined by a distinct lipid composition, as they move up and down the Golgi stack. Cargo can be exported to the plasma membrane from within the export domain found within every cisterna. Simulation and experimental testing of this rapid partitioning model produced all the key characteristics of the Golgi apparatus, including polarized lipid and protein gradients, exponential cargo export kinetics, and cargo waves, thus representing a viable description of the mechanism of intra-Golgi transport.
Biogenesis and dynamics of unconventional organelles
We explored the dynamics of several organelles, including autophagosomes, primary cilia, and midbodies. Autophagosomes form during autophagy, which is a highly conserved, bulk degradation pathway that is also involved in turnover of large aggregates and organelles within cells. While emerging results have revealed the importance of autophagy in various biological and pathological processes, how this pathway operates is far from clear. We used various live cell–imaging and molecular-genetic approaches to investigate the membrane origin of autophagosomes and the signals that recruit substrates to this organelle. Our data revealed that the outer membrane of mitochondria serves as the membrane source during starvation-induced autophagy formation and maturation. Furthermore, ubiquitin modification acts as a targeting signal for delivery of small cytosolic proteins and larger organelles to autophagosomes.
The primary cilium is a chemosensory and mechanosensory organelle that receives signals (both chemical and mechanical) from the extracellular environment. We imaged live polarized epithelial cells and found that cilia of one cell can make direct contact (bridges), which can be stable over many hours, with cilia of adjacent or nearby cells. The bridges are composed of cilia from individual cells; that is, they are not a continuation from one cell to the next. Cilia can thus initiate and mediate cell-cell communication through direct contact.
The midbody is the central region of the narrow membrane bridge that connects two daughter cells during cytokinesis. Cleavage of the narrow membrane neck on either side of the midbody is the final step in cytokinesis. We used structured illumination and time-lapse microscopy to clarify midbody cleavage, focusing on the role of endosomal sorting complexes required for transport (ESCRT). We found that the upstream ESCRT-I subunit TSG101 localizes to two cortical rings adjacent to the midbody center. The ESCRT-III subunit CHMP4B recruits to these rings, but at late stages in abscission also co-localizes with the narrow constriction at the site of membrane cutting. Direct visualization of the kinetics of ESCRT recruitment revealed an ordered assembly mechanism. The midbody protein CEP55 arrives early in cytokinesis, followed by ESCRT-I (TSG101), ESCRT-III (CHMP4B), and the ESCRT disassembly factor VPS4. The arrival of ESCRT-III and VPS4 coincides in time and space with cell separation, consistent with a direct role for these proteins in membrane abscission.
Mitochondrial morphology and its regulation of cell cycle progression
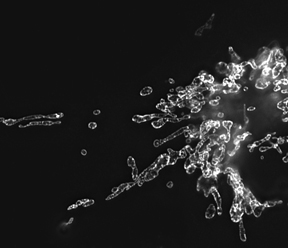
Mitochondria morphology in NRK cells
Image, obtained by structured illumination microscopy, of mitochondria labeled with GFP-tagged prohibitin.
Mitochondria are important energy-producing organelles within cells. Their morphology, including fragmented elements and tubular networks, results from a balance between fission and fusion events. To determine whether changes occur in mitochondrial dynamics at various stages of the cell cycle, we carried out live-cell imaging experiments in cells stably expressing RFP (red fluorescent protein) targeted to the mitochondrial matrix. We found that mitochondria exhibit distinct morphological and physiological states at different stages of the cell cycle. During mitosis, mitochondria fragment into hundreds of small units for partitioning into daughter cells at cytokinesis. Strikingly, at G1/S, mitochondria fuse into a single, huge, dynamic filamentous system, unlike at any other stage of the cell cycle. Photobleaching of an area across the filamentous system revealed that the mitochondrial matrix is continuous. In addition, the mitochondrial network is electrically coupled and has a higher membrane potential than mitochondria at any other stage of the cell cycle. When the filamentous network or its membrane potential was disrupted or its dynamics perturbed, cell cycle progression from G1 into S phase was arrested in a p53-dependent manner. Moreover, p21 overexpression, which induces a G1/S arrest, resulted in filamentous mitochondria with reduced matrix continuity and loss of electrical coupling. The data revealed that, during the cell cycle, mitochondrial dynamism and morphology undergo critical changes that are sensed by the cell at G1/S to control cell cycle progression.
Cytoskeletal and endomembrane cross-talk in polarized epithelial cells, hematopoietic niche cells, and the developing Drosophila syncytial blastoderm embryo
We studied cytoskeletal and endomembrane cross-talk between cells, using three cellular systems: polarized epithelial cells, hematopoietic niche cells, and developing Drosophila syncytial blastoderm embryos. Using monolayers of polarized MDCK cells, we employed live-cell imaging approaches to probe dynamin's role in integrating membranes and the cytoskeleton. Dynamin is a large GTPase, a regulatory mechano-enzyme that participates in nascent vesicle formation. Expression of the nucleotide-free form of dynamin led to dramatic apical constriction of expressing cells in the epithelial monolayer. The phenotype was dependent on cortactin, actin, and myosin II. General endocytic inhibitors and GTP-bound dynamin mutants did not cause apical constriction. Rather, the response was mediated by nucleotide-free/GDP-bound dynamin interacting with cortactin in an Erk kinase signaling pathway. Thus, in addition to its known roles in endocytosis, dynamin modulates the actomyosin contractile system.
Hematopoietic stem/progenitor cells (HSPC) reside in the bone marrow niche, where adhesive interactions with osteoblasts provide essential cues for the cells' proliferation and survival. We used live-cell imaging approaches to characterize the site of contact between osteoblasts and hematopoietic progenitor cells (HPC) as well as events at the contact site that result in downstream signaling responses important for niche maintenance. HPCs made prolonged contact with the osteoblast surface via a specialized membrane domain. At the contact site, the osteoblast took up portions of the specialized domain and internalized them into long-lived, SARA (Smad anchor for receptor activation)-positive, signaling endosomes, causing the osteoblasts to downregulate Smad signaling and increase their production of stromal-derived factor-1 (SDF-1), a chemokine responsible for HSPC homing to bone marrow. Targeted regulation of signaling and remodeling events within the osteoblastic niche microenvironment thus involves intercellular transfer—from HSPC to signaling endosomes within osteoblasts.
Patterning in the Drosophila embryo requires local activation and dynamics of proteins in the plasma membrane (PM). How these events are coordinated before cellularization—in the absence of PM barriers—remains unclear. We used in vivo fluorescence imaging to characterize the role of dynamin in this process. Dynamin was localized on short PM furrows during interphase, mediating endocytosis of PM components. Dynamin redistributed into the mitotic spindle region, off ingressed PM furrows, in metaphase, resulting in furrows with stabilized components. Preventing these cell cycle changes in dynamin distribution in a shibire fly mutant, in which dynamin is inactivated, resulted in dramatic morphogenetic consequences. During mitosis, these included dissociation from furrows of actin contractile machinery (including cortactin, diaphanous, anillin, and peanut), inhibition of furrow ingression, and aberrant spindle formation. During interphase, the consequences included randomization of surface proteins normally polarized to cap or intercap membranes and disruption of the diffusion barrier separating PM domains above nuclei. Based on these and other findings, we propose that cell cycle changes in dynamin GTPase activity orchestrate endocytic and actomyosin machinery for the regulation of PM furrows during early mitotic cycles in the fly embryo.
Additional Funding
- IATAP (Intramural AIDS Targeted Antiviral Program)
Publications
- Mitra K, Rassam B, Lin G, Lippincott-Schwartz J. A fused mitochondrial state with increased ATP production is linked to G1-S transition of the cell cycle. Proc Natl Acad Sci USA. 2009 106:1190-1195
- Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman C, Kanchanawong A, Davidson MW, Fetter RD, Hess HF. Single photon fluorescence interferometry for 3D super-resolution microscopy. Proc Natl Acad Sci USA. 2009 106:3125-3130
- Gillette J, Lippincott-Schwartz J. Intercellular transfer to signalling endosomes for targeted regulation within the hemapoietic stem cell-marrow niche. Nat Cell Biol. 2009 11:303-311
- Subach FV, Patterson GH, Renz M, Lippincott-Schwartz J, Verkhusha VV. Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. J Am Chem Soc. 2010 132:6481-6491
- Hailey DW, Peter K, Mitra K, Sougrat R, Lippincott-Schwartz J. Mitochondria supply membranes during the biogenesis of autophagosomes. Cell. 2010 141:656-667
- Lippincott-Schwartz J, Phair R. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu Rev Biophys. 2010 39:559-578
- Patterson G, Davidson M, Manley S, Hess H, Lippincott-Schwartz J. Super-resolution imaging using single molecule localization. Annu Rev Phys Chem. 2010 61:345-367
- Emr, S, Glick BS, Linstedt AD, Lippincott-Schwartz J, Luini a, Malhotra V, Marsh, BJ, Nakano A, Pfeffer SR, Rabouille C, Rothman JE, Warren G, Wieland FT. Journeys through the Golgi—taking stock in a new era. J Cell Biol. 2010 16:449-453
- Lippincott-Schwartz J, Patterson G. Imaging with photoactivatable fluorescent proteins. Trends Cell Biol. 2009 19:555-565
- Elia N, Lippincott-Schwartz J. Culturing MDCK cells in three dimensions for analyzing intracellular dynamics. Curr Protoc Cell Biol. 2009 4:4.22
- Kanodia JS, Rikhy R, Kim Y, Lund VK, DeLotto R, Lippincott-Schwartz J, Shvartsman SY. Dynamics of the Dorsal morphogen gradient. Proc Natl Acad Sci USA. 2009 106:21707-21712
- Mitra K, Lippincott-Schwartz J. Analysis of mitochondrial dynamics and functions using imaging approaches. Curr Protoc Cell Biol. 2010 4.25:1-21
Collaborators
- Win Arias, MD, Cell Biology and Metabolism Program, NICHD, Bethesda, MD
- Michael Davidson, PhD, Florida State University, Tallahassee, FL
- Robert Delotto, PhD, Københavns Universitet, Copenhagen, Denmark
- Harald Hess, PhD, Howard Hughes Medical Institute, Janelia Farm Campus, Ashburn, VA
- Robert Phair, PhD, BioInformatics, Rockville, MD
- Vladimir Verkhusha, PhD, Albert Einstein School of Medicine, Bronx, New York
- Stanislav Shvartsman, PhD, Princeton University, Princeton, NJ
Contact
For more information, email lippincj@mail.nih.gov or visit lippincottschwartzlab.nichd.nih.gov.


