You are here: Home > Unit on Microbial Pathogenesis
Host Cell Modulation by the Intracellular Pathogen Legionella pneumophila

- Matthias Machner, PhD, Head, Unit on Microbial Pathogenesis
- Maria Ramona Aldea, PhD, Postdoctoral Fellow
- Yang Chen, BA, Graduate Student
- Andrew Gaspar, PhD, Postdoctoral Fellow
- Antonia Thomas, BA, Summer Internship Student
Many Gram-negative bacteria use type IV secretion systems (T4SSs) to deliver bacterial effector proteins into host cells, where they modulate signaling events to create an environment supportive of bacterial growth.Legionella pneumophilais an opportunistic bacterial pathogen that, in humans, causes a life-threatening pneumonia called Legionnaires' disease. Upon phagocytosis by alveolar macrophages, the Dot/Icm T4SS delivers a large number of bacterial effector proteins into the host-cell cytosol (Figure 1). These effectors allow Legionella pneumophila to intercept proteins and membrane material of the infected cell, thereby transforming the Legionella-containing vacuole (LCV) into a protective compartment resembling host cell endoplasmic reticulum (ER). Legionella pneumophila mutants with a non-functional T4SS are avirulent, underscoring the importance of the translocated effector proteins for Legionella pneumophila infection. Our main focus is to identify and characterize host-pathogen interactions during Legionnaires' disease and to determine their importance for Legionella pneumophila virulence.
Effector proteins targeting vesicle transport GTPases
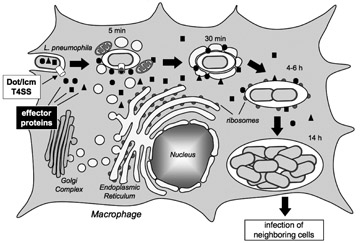
Figure 1. Legionella pneumophila intracellular replication cycle
L. pneumophila uses the Dot/Icm T4SS (white box) to deliver a multitude of effector proteins (black box) into the cytosol of an infected macrophage. The pathogen recruits host cell transport vesicles to establish a replication vacuole. (click image to enlarge)
We previously described two effector proteins from Legionella pneumophila, SidM and LidA, that target early secretory vesicle transport between the ER and the Golgi complex. These proteins modulate the activity of the small GTPase Rab1, the key regulator of this pathway. SidM mediates the recruitment of Rab1 to the Legionella-containing vacuole by combining two activities: it triggers release of Rab1 from its chaperone GDI (GDP dissociation inhibitor), an activity known as as GDI displacement factor (GDF) activity, and activates Rab1 by mediating exchange of GDP for GTP. Activated GTP-Rab1 is then released by SidM and most likely captured by LidA. The function of LidA and the molecular details of secretory vesicle binding to and fusing with the LCV are unclear, but several data indicate their importance for L. pneumophila replication vacuole formation. First, perturbations in early secretory vesicle trafficking reduced Legionella pneumophila intracellular replication. Second, L. pneumophila mutants deficient in SidM and LidA were defective for Rab1 accumulation and attenuated in intracellular survival. Third, the lidA allele is conserved in the genomes of all pathogenic L. pneumophila isolates analyzed to date. Together, these observations indicate that interception of Rab1-controlled secretory vesicle traffic by SidM and LidA plays a critical role in L. pneumophila virulence.
Interaction between the Legionella pneumophila effector protein LidA and the host cell GTPase Rab1
Unlike SidM whose function has been characterized in great detail, the role of LidA during Rab1 recruitment and vesicle hijacking is not understood. Using purified recombinant proteins, we found that LidA bound to active GTP-bound Rab1 as well as to Rab1 in its inactive GDP-loaded conformation. Gel permeation chromatography studies and analytical ultracentrifugation revealed that LidA and Rab1 formed a 1:1 complex. Binding was mediated through the central region of LidA enriched in coiled-coils, a typical Rab GTPase binding motif commonly found in eukaryotic proteins. LidA binding, however, did not alter the nucleotide binding state of Rab1, suggesting that LidA is a downstream effector of Rab1 rather than an upstream regulator. LidA binding to Rab1 prevented nucleotide extraction from Rab1 by EDTA, suggesting that LidA bound to Rab1 close to or on top of the nucleotide binding pocket. This tight interaction prevented cellular Rab1 ligands from interacting with Rab1, suggesting that LidA is not directly involved in processes that lead to the docking of secretory vesicles to the LCV.
Role of host cell Rab6 for Legionella pneumophila infection
Another host-cell GTPase targeted by LidA is Rab6, the regulator of retrograde vesicle transport from early/recycling endosomes to the trans-Golgi network and from the Golgi compartment to the ER. Using purified recombinant proteins in pulldown studies, we found that LidA preferentially bound to the active GTP-loaded form of Rab6. Similar to Rab1, Rab6 binding was mediated through the central region of LidA. Gel permeation chromatography revealed that LidA formed a 1:2 complex with Rab6, in contrast to the previously observed 1:1 stoichiometry described for the LidA–Rab1 complex. LidA binding efficiently inhibited inactivation of GTP-Rab6 by host-cell GTPase-activating protein (GAP), suggesting a role for this effector in maintaining an active pool of Rab6 within the infected host cell. Consistent with this observation, we found that overproduction of dominant inactive Rab6 interfered with Legionella pneumophila replication within COS1 cells, confirming the importance of Rab6-regulated processes for Legionella pneumophila infection.
Publications
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo Z-Q, O'Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. The E-Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2010; 29: In Press.
Collaborators
- Alfred L. Yergey, PhD, Biomedical Mass Spectrometry Facility, NICHD, Bethesda, MD
- Peter Backlund, PhD, Biomedical Mass Spectrometry Facility, NICHD, Bethesda, MD
Contact
For more information, email machnerm@mail.nih.gov or visit cbmp.nichd.nih.gov/ump/research.html


