You are here: Home > Section on Environmental Gene Regulation
Small Regulatory RNAs and Small Proteins

- Gisela Storz, PhD, Head, Section on Environmental Gene Regulation
- Aixia Zhang, PhD, Staff Scientist
- Cynthia M. Sharma, PhD, Visiting Scientist
- Fanette Fontaine, PhD, Visiting Fellow
- Chase L. Beisel, PhD, Postdoctoral Fellow
- Ryan T. Fuchs, PhD, Postdoctoral Fellow
- Errett C. Hobbs, PhD, Postdoctoral Fellow
- Lauren Waters, PhD, Postdoctoral Fellow
- Melissa Sandoval, BS, Postbaccalaureate Student
- Maureen K. Thomason, MS, Graduate Student
- Xuefeng Yin, BS, Graduate Student
Currently, we have two main interests: the identification and characterization of small noncoding RNAs and the identification and characterization of small proteins of less than 50 amino acids. For two reasons, small RNAs and small proteins have been overlooked. First, biochemical assays do not detect these small molecules. Second, genome annotation misses the corresponding genes, which are poor targets for genetic approaches. However, mounting evidence suggests that both classes of these small molecules play important regulatory roles.
Identification and characterization of small regulatory RNAs
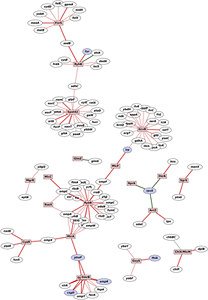
Click image to enlarge.
Figure 1. Networks regulated by trans-encoded base-pairing sRNAs in E. coli
The network is based on a compilation of published interactions. Boxes represent sRNAs and ovals represent mRNA targets. Ovals corresponding to mRNAs encoding transcription regulators are in blue. Red lines indicate negative regulation, green lines indicate positive regulation, and gray lines indicate base pairing but no effect on the target. The thickness of the lines indicates the level of proof for base pairing; compensatory mutation analysis supports those interactions represented with the thickest lines. A limited number of lines emanating from an sRNA may reflect incomplete characterization or the fact that the sRNA has only few targets.
During the past ten years, we have carried out a variety of systematic screens for small regulatory RNA genes in Escherichia coli. These screens, which have included computational screens for conservation of intergenic regions and direct detection after size selection or co-immunoprecipitation with the RNA binding protein Hfq, are all applicable to other organisms. We are now examining small RNA expression with tiled microarrays together with deep sequencing to extend further our identification of small RNAs, particularly antisense RNAs.
We focus on elucidating the functions of the small RNAs in E. coli. Early on, we showed that the OxyS RNA, whose expression is induced in response to oxidative stress, represses translation through limited base pairing with target mRNAs. We also discovered that the OxyS action is dependent on the Sm-like Hfq protein, which functions as a chaperone to facilitate OxyS RNA base pairing with its target mRNAs.
It is now clear that Hfq-binding small RNAs, which act through limited base pairing, are integral to many stress responses in E. coli (Figure 1). We found that the Hfq-binding RNA MicC, whose expression is induced in minimal medium and at low temperature, represses translation of the OmpC outer membrane porin. We also reported that the Hfq-binding small RNA FnrS, whose expression is induced by FNR upon a shift from aerobic to anaerobic conditions, down-regulates the levels of a variety of mRNAs encoding metabolic enzymes. The results suggest that the FnrS RNA extends the FNR regulon and increases the efficiency of anaerobic metabolism by repressing the synthesis of enzymes not needed under these conditions.
We recently showed that the Hfq-binding RNA Spot 42 plays a broad role in catabolite repression in E. coli by directly repressing genes involved in central and secondary metabolism, redox balancing, and the consumption of diverse non-preferred carbon sources. Many of the genes repressed by Spot 42 are transcriptionally activated by the global regulator CRP. Given that CRP represses Spot 42, these regulators participate in a specific regulatory circuit called a multioutput feedforward loop. We found that the loop can reduce leaky expression of target genes in the presence of glucose and can maintain repression of target genes under changing nutrient conditions. Our results suggest that base-pairing RNAs in feedforward loops can help shape the steady-state levels and dynamics of gene expression.
In addition to studying small RNAs that act through limited base pairing, we have become interested in small antisense RNAs that have the potential to form extensive base-pairing interactions with their mRNA targets encoded on the opposite strand. We showed earlier that base pairing between the GadY RNA and the 3′-untranslated region of the gadX mRNA encoded opposite GadY leads to increased levels of the gadX mRNA and GadX protein. Recently, we demonstrated that gadX is transcribed in an operon with gadW and that base pairing of GadY with the gadXW mRNA results in processing that gives rise to two halves accumulating at higher levels than the full-length mRNA. Interestingly, several enzymes, including the double-strand RNA–specific endoribonuclease RNase III, are involved in the GadY-direct cleavage.
We also found that a large class of antisense RNAs repress the synthesis of small toxic proteins. For example, in characterizing the Sib RNAs, which are encoded by five repeats in E. coli K-12, we observed an overexpression phenotype reminiscent of plasmid addiction. Further examination of the SIB repeat sequences revealed conserved open reading frames encoding highly hydrophobic 18–19 amino acid proteins (Ibs) opposite each sib gene. The Ibs proteins were toxic when overexpressed; such toxicity could be prevented by co-expression of the corresponding Sib RNA. Two other RNAs encoded divergently in another intergenic region were similarly found to encode a small hydrophobic protein (ShoB) and an antisense RNA regulator (OhsC). Computational screens together with experimental validation have shown that these small hydrophobic protein-antisense RNA gene modules, termed type 1 toxin-antitoxin modules, are much more widely distributed among bacteria than previously appreciated.
Studies are under way to characterize further other Hfq-binding RNAs and antisense RNAs and to elucidate the roles of small RNAs that act in ways other than base pairing.
Identification and characterization of small proteins
In our genome-wide screens for small RNAs, we found that several short RNAs encode small proteins. The correct annotation of the smallest proteins is one of the biggest challenges of genome annotation, and, perhaps more importantly, few annotated short open reading frames (ORFs) have been confirmed to correspond to synthesized proteins. Though these proteins have largely been missed, the few small proteins that have been studied in detail in bacterial and mammalian cells have been shown to have important functions in both signaling and cellular defenses. Thus, we initiated a project to identify and characterize E. coli proteins of fewer than 50 amino acids.
We used sequence conservation and ribosome-binding site models to predict genes encoding small proteins, defined as proteins of 16–50 amino acids, in the intergenic regions of the E. coli genome. We tested expression of the predicted and previously annotated small proteins by integrating a sequential peptide affinity tag directly upstream of the stop codon on the chromosome and assaying for synthesis by using immunoblot assays. This approach confirmed that 20 previously annotated and 18 newly discovered proteins of 16–50 amino acids are synthesized.
Remarkably, more than half of the newly discovered proteins are predicted to be single-transmembrane proteins, a prediction that prompted us to examine the localization, topology, and membrane insertion of the small proteins. Biochemical fractionation showed that, consistent with the predicted transmembrane helix, the small proteins are generally most abundant in the inner membrane fraction. We found examples of both Nin-Cout and Nout-Cin orientations in assays of topology-reporter fusions to representative small transmembrane proteins. Interestingly however, three of nine tested proteins displayed dual topology. Positive residues close to the transmembrane domains were conserved, and mutational analysis of one small protein, YohP, showed that the positive inside rule applied for single-transmembrane–domain proteins, as observed for larger proteins. Finally, fractionation analysis of small-protein localization in strains depleted of the Sec or YidC membrane insertion pathways uncovered differential requirements. Some small proteins appeared to be affected by both Sec and YidC depletion, others showed more dependence on one or the other insertion pathway, and one protein was not affected by depletion of either Sec or YidC. Thus, despite their diminutive size, small proteins display considerable diversity in topology, biochemical features, and insertion pathways.
We now are employing many of the approaches used by the group to characterize the functions of small regulatory RNAs in order to elucidate the functions of small proteins. Systematic assays for the accumulation of tagged versions of the proteins showed that many small proteins accumulate under specific growth conditions or after exposure to stress. We also generated and screened bar-coded null mutants and identified small proteins required for resistance to cell envelope stress and acid shock. In addition, we are exploiting the attached sequential peptide affinity tag to identify co-purifying complexes. The combination of approaches is yielding insights into when, where, and how the small proteins act. For example, we recently showed that expression of a 42–amino acid protein, now denoted MntS (formerly the small RNA gene rybA), is repressed by manganese through MntR. Overproduction of MntS causes manganese sensitivity, while a lack of MntS perturbs proper manganese-dependent repression of another manganese-regulated gene. Drawing on these results, we propose that MntS plays a novel role in intra-cellular manganese trafficking and homeostasis.
Additional Funding
- PRAT Fellowship Award to Dr. Lauren Waters (2009-2011)
- Life Sciences Research Foundation to Dr. Chase Beisel (2010-2012)
Publications
- Opdyke JA, Fozo EM, Hemm MR, Storz G. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol 2011;406:29-43.
- Beisel CL, Storz G. The base pairing RNA Spot 42 participates in a multi-output feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell 2011;41:286-297.
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 2011;43:880-891.
- Hobbs EC, Fontaine F, Yin X, Storz G. An expanding universe of small proteins. Curr Opin Microbiol 2011;14:167-173.
- Fontaine F, Fuchs RJ, Storz G. Membrane localization of small proteins in Escherichia coli. J Biol Chem 2011;286:32464-32474.
Collaborators
- Susan Gottesman, PhD, Laboratory of Molecular Biology, NCI, Bethesda, MD
- Jörg Vogel, PhD, University of Würzburg, Germany
- Karen M. Wassarman, PhD, University of Wisconsin, Madison, WI
Contact
For more information, email storz@helix.nih.gov or visit cbmp.nichd.nih.gov/segr.

