You are here: Home > Section on Neuronal Connectivity
Assembly and Function of Drosophila Visual Circuits

- Chi-Hon Lee, MD, PhD, Head, Section on Neuronal Connectivity
- Chun-Yuan Ting, PhD, Staff Scientist
- Thangavel Karuppudurai, PhD, Postdoctoral Fellow
- Yang Lee, PhD, Postdoctoral Fellow
- Tzu-Yang Lin, PhD, Postdoctoral Fellow
- Krishna Melnattur, PhD, Postdoctoral Fellow
- Moyi Li, BA, Biological Laboratory Technician
Using the Drosophila visual system as a model, we study how neurons form complex yet stereotyped synaptic connections during development and how the assembled neural circuits extract visual attributes, such as color and motion, to guide animal behaviors. Flies' vision is mediated by three types of photoreceptor, R1-6, R7, and R8, which each respond to a specific spectrum of light. The three types of photoreceptor project their axons to three distinct layers in the brain, where the cells form synaptic connections with different target neurons. To study visual circuit functions, we combined structural and functional approaches to map visual circuits. Using both light- and electron-microscopy studies, we identified the medulla neurons that are post-synaptic to R7 and R8 photoreceptors. We determined their usage of neurotransmitters and receptors by single-cell transcript profiling. By systematically "inactivating" or "restoring" the functions of specific neuronal types and examining behavioral consequences, we are determining the role of these medulla neurons in processing color information. For circuit development, we focus on the formation of synaptic circuits between medulla neurons and R7 and R8 photoreceptors. We use high-resolution imaging techniques and genetic manipulations to determine the molecular mechanisms that control dendritic patterning and synaptic specificity of the medulla neurons.
Mapping color-vision circuits
Color vision, which distinguishes spectral compositions independent of brightness, provides animals—from insects to primates—with great power for object recognition and memory registration and retrieval. Using a combination of genetic, histological, electrophysiological, and behavioral approaches, we study how the visual system processes chromatic information to generate color perception in Drosophila. Previous primate studies suggest that the color opponent neurons, which exhibit combination-sensitive excitatory and/or inhibitory interaction between two or more photoreceptor types, subserve color vision. Our anatomical study suggests that the Drosophila color-vision circuit in the medulla likely contains such color opponent neurons. We hypothesize that chromatic information is processed in two discrete stages in Drosophila. First, the transmedulla (Tm) neurons serve as color opponent neurons by combining multiple photoreceptor channels and relaying the information to the higher visual center, the lobula. Second, chromatic information is spatially integrated in the lobula, to subserve higher-order color-vision functions, such as color contrast and constancy. Using color preference and color learning assays, we demonstrated that flies innately prefer short wavelengths of light but they can be trained to select specific wavelengths of light by Pavlovian conditioning, indicating that flies, like honeybees and humans, have true color vision.
Using both light and electron microscopy, we determined the synaptic circuits of photoreceptors and their synaptic target neurons in the medulla. The chromatic photoreceptors—R7 (UV–sensing) and R8 (blue/green-sensing)—provide inputs to a subset of first-order interneurons, which might serve as color opponent neurons. The first-order interneurons Tm5a, Tm5b, and Tm5c receive direct synaptic inputs from R7 while Tm9, Tm20, and Tm5c receive inputs from R8. In addition, the Tm neurons receive indirect inputs from R1-6 via L3 and relay spectral information from the medulla to the higher visual center, the lobula. In addition to the direct pathways from photoreceptors to Tm neurons, the amacrine neuron Dm8 receives input from multiple R7s and provides input for Tm5a, Tm5b, and Tm5c.
To relate neural connectivity to functions, it is critical to assign components of synaptic machinery to specific connections. To directly probe which neurotransmitters and receptors determine the polarity and dynamics of signal transmission, we developed a method to profile transcripts in single neurons. We used highly specific promoter–Gal4 constructs to label single types of neurons with GFP, isolated these GFP–labeled neurons from adult fly brains, and profiled their gene expression patterns by RT–PCR. Using this method, we determined that the majority of the first-order interneurons in the chromatic circuits express the vesicular glutamate transporter and the kainate-type of ionotropic glutamate receptor, indicating that the neurons provide and receive fast, sign-conserving glutamate inputs.
Our previous studies revealed that amacrine Dm8 neurons are both required and sufficient for animals' innate spectral preference to UV light while Tm9 neurons are sufficient to drive green phototaxis. RNAi-knockdown of the vesicular glutamate transporter in Dm8 significantly reduced UV preference, suggesting that glutamatergic output of Dm8 is critical for this neuron's functions. Furthermore, Tm5, a neural type downstream of Dm8, is functionally required for optimal UV preference—inhibiting the synaptic transmission of Tm5 significantly reduced animals' attraction to UV over green light. Taken together, our results suggest that Tm5 neurons receive excitatory glutamate input from Dm8 neurons to mediate UV phototaxis.
Dendritic development of Drosophila optic lobe neurons
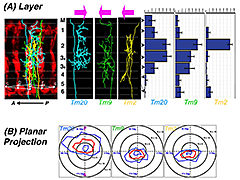
Click image to enlarge.
Figure 1. Type-specific dendritic attributes
Three classes of medulla neurons, Tm20 (colored cyan), Tm9 (green), Tm2 (yellow), route their dendrites to specific layers (A) in distinct planar directions (B). Left upper panels: the neuronal processes of three neuronal classes were imaged by confocal microscopy, traced, and standardized by non-linear registration. A: anterior; P: posterior. Medulla layers (M1-M6) are as indicated (left middle panel). Histograms show the layer distribution of terminal nodes for each class (right upper panel). Each neuronal class project dendrites in distinct planar directions as shown in the polar plots (lower panels). Means and standard deviations are colored in blue and red, respectively.
Our long-term goal is to determine how visual circuits are assembled during development. Most regions of vertebrate and invertebrate brains are organized in columns and layers, which facilitate information processing and propagation. During development, axons and dendrites are routed to specific layers and columns. Spatial coincidence of axons and dendrites affords a key specificity determinant for synaptic pairing. While much has been discovered about axonal guidance in the past years, essentially nothing is known about how target neurons extend dendrites in type-specific patterns to pair with pre-synaptic terminals of afferents. Much of our current understandings of dendrite development came from the studies of Drosophila Da neurons in the peripheral nervous system, which extend dendrites not to specific targets but to populate (or tiling) a two-dimensional receptive field. Two key questions concerning dendritic development remain largely unanswered, namely, how CNS neurons establish type-specific dendritic arborization patterns in three-dimensional space and how synaptic partnership between axons and dendrites are matched.
Using Drosophila optic lobe neurons as a model, we aim to determine the molecular mechanisms by which transmedulla (Tm) neurons extend dendrites to specific layers and columns to match with presynaptic terminals of photoreceptors. Like the vertebrate cortex and retina, the medulla neuropil is organized in columns and layers, suggesting that fly medulla neurons and vertebrate cortex neurons confront similar challenges in routing their dendrites to specific layers and columns. In addition, the fly visual system has several unique advantages: (i) the medulla neurons extend dendritic arbors in a three-dimensional lattice structure, facilitating morphometric analysis; (ii) the presynaptic targets for many medulla neuron types are known from our anatomical studies; (iii) our group has developed genetic tools for labeling specific classes of medulla neurons and determining their connectivity.
We developed several techniques to analyze dendritic structures in three-dimensional space. First, to image reliably the slender dendrites of medulla neurons, we developed a dual-view imaging technique that generates isotropic 3D-images of dendrites at the diffraction limit. In combination with super-resolution techniques, such as the time-gated STED (stimulated-emission depletion) nanoscope, the image quality can be further improved. Second, we developed an image-registration technique that makes use of the regular array structures of the optic lobe to standardize dendritic branching patterns by non-linear registration. In combination with a series of statistical methods we established, the technique allows us to analyze dendritic patterns in a standard 3D-lattice space. Third, we developed a single-cell split–GFP system which, in combination with a pre-synaptic marking technique, detects synaptic contacts between two classes of neurons at the light-microscopic level. We generated a data set of three types of medulla neurons. Our analyses reveal that, within each class of neurons, the axonal positions and dendritic projection patterns are stereotypic whereas the detailed branching topology is not conserved. Furthermore, while layer- and planar-specific routing of dendrites and axonal positions can be used to differentiate neuronal types, layer-specific dendritic routing provides the strongest classifier.
To determine the molecular mechanisms that govern dendritic routing, we took a candidate approach and examined a series of available mutants. Our previous study revealed that R7 and R8 photoreceptor axons express TGF-beta/activin, which could potentially signal the medulla Tm neurons in an antegrade fashion, in addition to its known autocrine-signaling role in R7s. We found that TGF-beta signaling is required in two R7/8 synaptic target neurons, Dm8 and Tm20, for appropriate dendritic patterning. Using the imaging and statistical analysis tools we developed, we characterized the loss-of-function mutant phenotypes and found that the mutant Dm8 and Tm20 neurons devoid of TGF-beta signaling expanded larger but less complex dendritic trees. Segment analyses further revealed that both dendritic branching and terminating frequencies were reduced in the mutant neurons. Using the single-cell Split-GFP method, we detected a corresponding increase in synaptic contacts between the Tm20 dendrites and R8 photoreceptor axons. Taken together, our results demonstrated that TGF-beta signaling regulates dendritic patterning of Tm neurons and consequentially their synaptic partnership with photoreceptors.
Publications
- Ting C-Y, Gu S, Guttikonda S, Lin T-Y, White BH, Lee C-H. Focusing transgene expression in Drosophila by coupling Gal4 with a novel split-LexA expression system. Genetics 2011;188:229-233.
- Brody T, Yavatkar AS, Kuzin A, Tyson L, Kundu M, Ross J, Lin T-Z, Lee C-H, Awasaki T, Lee T, Odenwald WF. Use of a Drosophila genome-wide conserved sequence database to identify functionally related cis-regulatory enhancers. Dev Dyn 2011;241:169-189.
- Takemura S, Karuppudurai T, Ting C-Y, Lu Z, Lee, C-H, Meinertzhagen I. Cholinergic circuits integrate neighboring visual signals in a Drosophila motion detection pathway. Curr Biol 2011;21:2000-2005.
- Wardill TJ, List O, Li X, Dongre S, McCulloch M, Ting C-Y, O'Kane CJ, Tang S, Lee C-H, Hardie RC, Juusola M. Multiple spectral inputs contribute to motion discrimination in the Drosophila visual system. Science 2012;336:925-931.
- Meinertzhagen IA, Lee C-H. The genetic analysis of functional connectomics in Drosophila. Adv Genet 2012;in press.
Collaborators
- Matthew McAuliffe, PhD, Division of Biomedical Imaging Research Services Section, CIT, NIH, Bethesda, MD
- Philip McQueen, PhD, Mathematical and Statistical Computing Laboratory, CIT, NIH, Bethesda, MD
- Ian Meinertzhagen, PhD, DSc, Dalhousie University, Halifax, Canada
- Nishith Pandya, BA, Division of Biomedical Imaging Research Services Section, CIT, NIH, Bethesda, MD
- Thomas Pohida, MSEE, Division of Computational Bioscience, CIT, NIH, Bethesda, MD
- Randy Pursley, MSEE, Division of Computational Bioscience, CIT, NIH, Bethesda, MD
- Mark Stopfer, PhD, Program in Developmental Neuroscience, NICHD, Bethesda, MD
- Benjamin White, PhD, Laboratory of Molecular Biology, NIMH, Bethesda, MD

