You are here: Home > Section on Nutrient Control of Gene Expression
Transcriptional and Translational Regulatory Mechanisms in Nutrient Control of Gene Expression

- Alan G. Hinnebusch, PhD, Head, Section on Nutrient Control of Gene Expression
- Hongfang Qiu, PhD, Staff Scientist
- Neelam Sen, PhD, Research Fellow
- Sebastien Lagiex, PhD, Postdoctoral Fellow
- Pilar Martin-Marcos, PhD, Postdoctoral Fellow
- Eun-Hee Park, PhD, Postdoctoral Fellow
- Attiq Rehman, PhD, Postdoctoral Fellow
- Adesh Saini, PhD, Postdoctoral Fellow
- Jyothsna Visweswaraiah, PhD, Postdoctoral Fellow
- David Young, PhD, Postdoctoral Fellow
- Fujun Zhou, PhD, Postdoctoral Fellow
- Jinsheng Dong, PhD, Senior Research Assistant
- Fan Zhang, MS, Senior Research Assistant
- Cuihua Hu, BA, Research Assistant
- Nazanin Ashourian, BS, Graduate Student
We study molecular mechanisms of gene regulation at the translational and transcriptional levels, using the regulation of amino acid–biosynthetic genes in budding yeast as a model system. Transcription of these and many other genes is coordinately induced by the transcriptional activator Gcn4 in response to starvation of any amino acid. Expression of GCN4 is coupled to amino acid levels by a conserved translational control mechanism involving upstream open reading frames (uORFs) in GCN4 mRNA. Ribosomes translate the 5′-most uORF (uORF1) and, under non-starvation conditions, reinitiate translation at uORFs 2, 3, or 4 and then dissociate from the mRNA, keeping GCN4 translation repressed. In starvation conditions, the reinitiating ribosomes bypass uORFs 2–4 and reinitiate at GCN4 instead, owing to decreased availability of the ternary complex (TC)—comprised of initiation factor 2 (eIF2), GTP, and initiator Met-tRNAi—which binds to the small (40S) ribosomal subunit to assemble a 43S preinitiation complex (PIC). TC abundance is reduced in starved cells by phosphorylation of the alpha subunit of eIF2 (eIF2a) by Gcn2, a protein kinase conserved in all eukaryotes, converting eIF2 from substrate to inhibitor of its guanine nucleotide exchange factor (GEF) eIF2B. Hence, GCN4 translation is an in vivo indicator of impaired TC loading on 40S subunits. We previously exploited this fact to isolate mutations in subunits of eIF2B that constitutively derepress GCN4 (Gcd− phenotype) by lowering TC assembly in the absence of eIF2 phosphorylation. More recently, we used the Gcd− selection to identify domains/residues in eIF1, eIF1A, and eIF3, and also residues of 18S rRNA located near the "P" decoding site of the 40S subunit, that participate in rapid TC recruitment in vivo. In collaboration with Jon Lorsch's group, we demonstrated that segments/residues in eIF1, eIF1A, and 18S rRNA, which are implicated genetically in TC recruitment, also stimulate this reaction in a fully reconstituted in vitro system.
We also investigate the roles of various eIFs and the 40S subunit in scanning the mRNA 5′ untranslated region and accurately identifying the AUG initiation codon. The studies exploit a genetic selection for mutations that elevate initiation at near-cognate UUG start codons (Sui− phenotype) or suppress this aberrant initiation event (Ssu− phenotype). In this way, we provided the first evidence that eIF1A and the c-subunit of eIF3 make critical contributions to accurate AUG recognition, and we localized these functions to the unstructured N- and C-tails of eIF1A and the N-terminal domain of eIF3c. In collaboration with Lorsch's group, we also provided strong evidence that dissociation of eIF1 from the PIC is a critical step in AUG recognition, indicating that this factor serves as a "gate-keeper" necessary to suppress initiation at non-AUG triplets. Biochemical analysis revealed that eIF1 stabilizes an "open" conformation of the 40S subunit conducive to scanning and loading of the TC in a metastable state, which enables inspection of successive triplets as they enter the P site for complementarity with the anticodon of Met-tRNAi. However, eIF1 must dissociate from the PIC for AUG selection, as the absence of eIF1 favors the closed, scanning-arrested conformation of the 40S subunit to which the TC is more tightly bound. Our recent analysis of eIF1A provided strong evidence that 10–amino acid repeats in the C-terminal tail (dubbed "scanning enhancers", or SE elements) function together with eIF1 to stabilize the open complex and promote the first step of TC loading, but then must be ejected from the P site to achieve rearrangement to the closed complex, with TC more stably bound and Met-tRNAi base-paired with AUG. Furthermore, we identified scanning-inhibitory (SI) elements in the N-terminal tail and helical domain of eIF1A that oppose the SE elements and promote rearrangement to the closed complex necessary for AUG recognition.
In the arena of transcriptional control, we previously identified multiple co-activators required for gene activation by Gcn4 and defined the molecular program for recruitment of nucleosome-remodeling enzymes and adaptor proteins that remove repressive chromatin structure and recruit TATA–binding protein and other general transcription factors, along with RNA polymerase II (Pol II) to Gcn4 target promoters. We also demonstrated the co-transcriptional recruitment of the histone acetyltransferase (HAT) complexes SAGA and NuA4 to transcribed coding sequences and obtained evidence that they cooperate to promote histone eviction from transcribed coding sequences and to stimulate the rate of Pol II elongation in vivo. We further described a two-stage recruitment mechanism for NuA4, involving the serine-5 phosphorylated C-terminal domain (CTD) of Pol II and methylated histones, and more recently extended the mechanism to include the histone deacetylase (HDA) complex Rpd3C(S). In addition, we demonstrated that the HDAs Rpd3, Hos2, and Hda1 have overlapping functions in deacetylating coding-sequence nucleosomes and observed a strong correlation between increased acetylation and lower histone occupancy in various HDA mutants. The results support the notion that histone acetylation is a key determinant of nucleosome disassembly in transcribed coding sequences.
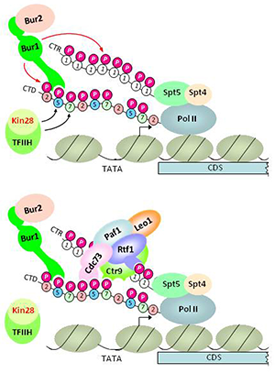
Pol II CTD kinases Bur1 and Kin28 promote an Spt5–independent pathway for recruitment of transcription elongation factor Paf1C.
Click image to enlarge.
Figure 1. Model for dual pathway of Paf1C recruitment by phosphorylated forms of Spt5 C-terminal repeats and the Pol II C-terminal domain
Upper panel: Kin28 phosphorylates Ser5 and Ser7 in the Pol II CTD, and the Ser5P stimulates Bur1/Bur2 recruitment to the CTD via the Bur1–phosporylated CTD/CTR interaction domain (PCID). Spt5/Spt4 associates with Pol II via the "clamp domain" of Pol II, independently of CTD phosphorylation. Recruited Bur1/Bur2 phosphorylates Ser2 (and possibly Ser7) in CTD repeats also harboring Ser5P to produce diphosphorylated repeats, and also phosphorylates the Spt5 CTRs (upper panel).
Lower panel: Paf1C recruitment occurs through cooperation between interactions of the PCIDs in Cdc73, Rtf1, and Ctr9 with diphosphorylated CTD repeats and phosphorylated Spt5 CTRs.
Previously, we showed that the cyclin-dependent kinase (Cdk) Bur1 is recruited to promoters through its direct interaction with the Ser5–phosphorylated Pol II CTD (generated by Kin28), phosphorylates Ser2 in CTD repeats near the promoter, and also stimulates Ser2–CTD phosphorylation at promoter-distal sites by the Cdk Ctk1. Recently, it was demonstrated that Bur1 also phosphorylates C-terminal repeats (CTRs) of elongation factor Spt5 in a manner that facilitates co-transcriptional recruitment of the elongation factor Paf1 complex (Paf1C), which promotes co-transcriptional histone H3 methylation of Lys4 and Lys35. However, the biochemical function of the phosphorylated Spt5 CTR in Paf1C recruitment was unknown. It was also unknown whether the phosphorylated Pol II CTD participates in Paf1C recruitment. Our recent studies indicate that native Paf1C and Paf1C subunit Cdc73 bind specifically to both phosphorylated Spt5 CTRs and Ser2,Ser5–diphosphorylated Pol II CTD repeats in vitro and that mutations eliminating these binding activities of Cdc73 reduce Paf1C recruitment in vivo. To establish that Kin28 and Bur1 promote Paf1C independently of the phosphorylated Spt5 CTRs, we generated phosphomimetic (aspartate) mutants of the Spt5 CTR and showed that they can sustain Paf1C recruitment in otherwise wild-type cells, but not following inactivation of an analog-sensitive variant of Bur1 in vivo. Inhibiting Kin28 reduces phosphorylation of both Spt5 and Ser2 of the CTD, consistent with Kin28 stimulation of Bur1 recruitment; however, inactivation of Kin28 also impairs Paf1C recruitment in the presence of the Spt5 phosphomimetic CTR mutant. Moreover, inactivating the CTD–interaction domain (CID) of Cdc73 further reduces histone H3 methylation of Lys-4 and Lys-36 in cells harboring nonphosphorylatable, Ala-substituted Spt5 CTRs. The results identify an Spt5–independent pathway of Paf1C recruitment and suggest that diphosphorylated CTD repeats and the Spt5 phospho–CTR mediate parallel pathways of Paf1C recruitment that both depend on Kin28 and Bur1 (Figure 1).
Coupling transcriptional activation by Gcn4 to endosome function
Previously, we reported that disruption of vesicular protein trafficking at the late endosome/MVB (multivesicular body), occurring in a subset of vps mutants, impedes the ability of nucleus-localized and UAS–bound Gcn4 to activate transcription. We recently collaborated with Vytas Bankaitis' group to establish that Gcn4 function is similarly down-regulated under conditions of sterol limitation, in a manner that depends on the sterol-binding protein Kes1. Overexpressed Kes1 was shown to bind to endomembranes and impede protein trafficking in the cytoplasm and to reduce the transcription of Gcn4 target genes in the nucleus. Cellular sphingolipids (SL) are elevated under these conditions, and it was found that addition of phytosphingosine (PHS) to cells impairs Gcn4 function in a KES1-dependent manner. ChIP analysis revealed that PHS reduces Pol II occupancy at ARG1 to an extent that exceeds the decline in UAS occupancy by Gcn4, suggesting a transcriptional defect of the type described previously in vps mutants. Interestingly, Mediator subunit Srb10 is required for the growth inhibition and impairment of Gcn4 function evoked by Kes1 overexpression or PHS addition. Bankaitis et al. propose that Kes1 provides a sterol-regulated "brake" on PI(4)P–directed vesicular protein transport and that increased Kes1 binding to endomembranes at low sterol levels evokes increased cellular SL levels as the result of aberrant expansion of the endosome, which in turn provokes an Srb10–dependent impairment of Gcn4 function in the nucleus.
Functional determinants in eIFs -1, -1A and -2b discriminate against both poor AUG context and non-AUG start codons.
In an effort to map functional domains in eIF1 involved in accurate start codon selection, we recently isolated Sui- and Ssu- mutations in eIF1 that, respectively, increase or decrease the frequency of translation initiation at near-cognate UUG codons. Interestingly, we found that the Sui- eIF1 proteins are overexpressed, while the Ssu- eIF1 mutants are relatively underexpressed, compared with wild-type eIF1. This suggested that eIF1 might negatively autoregulate translation by inhibiting recognition of its AUG codon, which occurs in a poor sequence context for initiation in yeast (C-3-G-2-U-1-AUG). Indeed, Atkins et al. recently showed that mammalian eIF1 autoregulates translation in this manner. We provided strong evidence that yeast eIF1 discriminates against poor AUG context, in addition to near-cognate initiation codons, by showing that the opposite effects of eIF1 Sui- and Ssu- mutations on eIF1 protein levels, and expression of a SUI1-lacZ fusion, were diminished by introducing the optimal context (A-3-A-2-A-1-AUG) at the (SUI1) start codon for eIF1 translation. Moreover, Sui- mutations in eIF2b (SUI3-2) or the eIF1A SE elements also suppress the negative effect of poor context on SUI1 expression, whereas the eIF1A Ssu- mutation 17-21 exacerbates poor context, as we observed for Sui- and Ssu- mutations in eIF1 itself. The fact that discrimination against weak AUG context involves the same residues in these eIFs that discriminate against non-AUG start codons suggests that the context and AUG act together to promote the transition from the open, scanning-conducive conformation of the PIC to the closed conformation for start codon recognition.
Interaction of the unstructured N-terminal tail of eIF2b with the eIF5 C-terminal domain is required for eIF1 dissocation from the 40S subunit and start codon recognition.
Previously, we showed that the CTD of eIF5 interacts with multiple initiation factors (eIFs 1, eIF3c, and eIF2b) to stabilize the multi-factor complex (MFC), now regarded as the form in which these factors initially bind to the 40S ribosomal subunit. In collaboration with Gerhard Wagner's and Jon Lorsch's groups, we obtained evidence that the eIF5-CTD/eIF2b-NTT interaction is crucial for start codon recognition. We identified the binding sites for mammalian eIF1 and eIF2b-NTT by NMR spectroscopy and found that they overlap on the same face of the eIF5-CTD and that their interactions with the eIF5-CTD are mutually exclusive. A double-lysine substitution ("KK") in the eIF5-CTD was found to selectively impair eIF5-CTD binding to eIF2b-NTT but not eIF1. We found that the KK substitution in yeast eIF5 has an Ssu- phenotype, suppressing the elevated UUG:AUG initiation ratio conferred by eIF1 Sui- substitutions in the 40S binding surface of eIF1, which provoke inappropriate eIF1 dissociation from the PIC at UUG start codons. By contrast, the eIF5 KK substitution does not suppress the elevated UUG:AUG initiation ratio produced by a different class of eIF1 Sui- substitions (G107R/K) that delay eIF1 dissociation specifically at AUG codons. In fact, the double mutant harboring the KK and G107R mutations displays a strong defect in bulk initiation at AUG codons. We showed previously that high concentrations of eIF5 can reverse the aberrant retention of the eIF1-G107K variant on the 40S subunit in reconstituted PICs and, importantly, that the KK substitution impairs this activity of eIF5. Together, these findings indicate that the eIF5-CTD/eIF2b-NTT interaction (impaired by the eIF5 KK substitution) is important for proper eIF1 dissociation at the start codon (Figure 2).
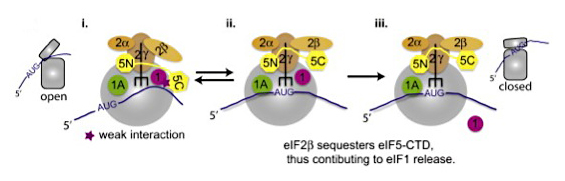
Figure 2. Schematic model depicting the contribution of the eIF5-CTD/eIF2b-NTT interaction in promoting the release of eIF1 on AUG start codon recognition
(i) The open 43S conformation allows for mRNA recruitment and relatively unstable binding of the eIF2-GTP-Met-tRNAi ternary complex. During the assembly stage of the PIC, the eIF5–CTD interacts with eIF1. (ii) The 43S PIC scans the mRNA in an open conformation until start codon recognition. During this scanning stage the eIF5–NTD induces eIF2γ to cleave GTP. Start codon recognition sets the stage for large conformational rearrangements on the subunit interface ensuring the closed state of the PIC. eIF2β exhibits a stronger affinity for the overlapping binding surface of the eIF5–CTD than eIF1; hence, the disruption of the eIF1:eIF5–CTD interaction by eIF2β allows for an indirect mechanism to dislodge eIF1 from the 43S PIC. Upon release of eIF1, the free phosphate is subsequently released. (iii) eIF5–CTD stabilizes the closed ribosomal conformation of the PIC on start codon selection.
Specific domains in eIF4G bias RNA unwinding activity of the eIF4F complex towards duplexes with 5′ overhangs.
Binding of eIF4F (eIF4E, eIF4G, eIF4A) to the mRNA cap structure recruits the helicase eIF4A to the 5′ UTR of mRNA, where it can promote 43S PIC attachment and scanning by unwinding secondary structures. In collaboration with Jon Lorsch's group, we obtained evidence that the three RNA–binding domains in the scaffold molecule eIF4G impart a preference in yeast eIF4A for unwinding duplexes with single-stranded 5′ overhangs, which should help to direct eIF4A to the 5′ UTR and provide 5′ to 3′ directionality to scanning.
Distinct roles of translation elongation factors 3 and 1A in modulating the function of eIF2a kinase Gcn2
Recycling of eIF2-GDP to eIF2-GTP by eIF2B is impaired under stress conditions, such as amino acid starvation, by phosphorylation of eIF2a by kinase Gcn2. The resulting reduction in TC abundance down-regulates general protein synthesis but specifically induces translation of transcription factor Gcn4. The kinase function of Gcn2 is activated on translating ribosomes by uncharged tRNA occupying the A-decoding site in a manner requiring Gcn2 interaction with Gcn1 bound to the same ribosomes. Translation requires cyclical association of eukaryotic elongation factors (eEFs) with the ribosome, and part of the ribosome-binding domain in Gcn1 has homology to eEF3, suggesting that these proteins use overlapping binding sites on the ribosome. Indeed, we found that overexpressing the ribosome-binding segments of eEF3 impedes eIF2a phosphorylation by Gcn2 in cells, consistent with eEF3 interference with Gcn1–regulatory function on the ribosome. Hence, we propose that the Gcn1–Gcn2 complex functions only on ribosomes with A-site–bound uncharged tRNA, because eEF3 does not occupy these stalled elongation complexes. We also obtained evidence that Gcn2 interacts with eEF1A in vivo in a manner independent of their separate ribosome interactions and that the Gcn2–eEF1A interaction is diminished in amino acid–starved cells. These findings suggested that eEF1A could function as a Gcn2 inhibitor; consistent with this possibility, we showed that purified eEF1A reduced the ability of Gcn2 to phosphorylate its substrate, eIF2α, but did not diminish Gcn2 autophosphorylation in vitro. Thus, it appears that both eEF1A and eEF3 negatively regulate Gcn2 activation on translating ribosomes by distinct mechanisms, limiting Gcn2 activation to stalled ribosomes harboring uncharged tRNA in amino acid-starved cells.
Additional Funding
- International Human Frontiers of Science Program Grant RGP0028
Publications
- Visweswaraiah J, Lageix S, Castilho BA, Izotova L, Kinzy TG, Hinnebusch AG, Sattlegger E. Evidence that eukaryotic translation elongation factor 1A (eEF1A) binds the Gcn2 protein C terminus and inhibits Gcn2 activity. J Biol Chem 2011;286:36568-36579.
- Martin-Marcos P, Cheung YN, Hinnebusch, AG. Functional elements in initiation factors 1, 1A, and 2beta discriminate against poor AIG context and non-AUG start codons. Mol Cell Biol 2011;31:4814-4831.
- Mousley CJ, Yuan P, Gaur NA, Trettin KD, Nile AH, Deminoff SJ, Dewar BJ, Wolpert M, Macdonald JM, Herman PK, Hinnebusch AG, Bankaitis VA. A sterol-binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell 2012;148:702-715.
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus MA, Akabayov B, Milbradt AG, Luna LE, Seo HC, Hyberts SG, Fahmy A, Reibarkh M, Miles D, Hagner PR, O'Day EM, Yi T, Marintchev A, Hinnebusch AG, Lorsch JR, Asano K, Wagner G. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with elF1 and elF2β. Cell Rep 2012;1:689-702.
- Qiu H, Hu C, Gaur NA, Hinnebusch AG. Poll II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J 2012;31:3494-3505.
Collaborators
- Katsura Asano, PhD, Kansas State University, Manhatten, Kansas, KS
- Vytas Bankaitis, PhD, University of North Carolina School of Medicine, Chapel Hill, NC
- Terri Kinzy, PhD, Robert Wood Johnson Medical School-University of Medicine and Dentistry of New Jersey, Piscataway, NJ
- Jon Lorsch, PhD, The Johns Hopkins University School of Medicine, Baltimore, MD
- Assen Marintchev, PhD, Boston University School of Medicine, Boston, MA
- Evelyn Sattlegger, PhD, Massey University, Auckland, New Zealand
- Gerhard Wagner, PhD, Harvard Medical School, Boston, MA
Contact
For more information, email hinnebua@mail.nih.gov or visit sncge.nichd.nih.gov.

