You are here: Home > Section on Nutrient Control of Gene Expression
Transcriptional and Translational Regulatory Mechanisms in Nutrient Control of Gene Expression

- Alan G. Hinnebusch, PhD, Head, Section on Nutrient Control of Gene Expression
- Hongfang Qiu, PhD, Staff Scientist
- Neelam Sen, PhD, Research Fellow
- Sebastien Lagiex, PhD, Postdoctoral Fellow
- Pilar Martin-Marcos, PhD, Postdoctoral Fellow
- Eun-Hee Park, PhD, Postdoctoral Fellow
- Jyothsna Visweswaraiah, PhD, Postdoctoral Fellow
- Fnu Yashpal, PhD, Postdoctoral Fellow
- David Young, PhD, Postdoctoral Fellow
- Quira Zeiden, PhD, Postdoctoral Fellow
- Fujun Zhou, PhD, Postdoctoral Fellow
- Jinsheng Dong, PhD, Senior Research Assistant
- Fan Zhang, MS, Senior Research Assistant
- Cuihua Hu, BA, Research Assistant
- Nazanin Ashourian, BS, Graduate Student
- Laura Marler, BS, Graduate Student
We study molecular mechanisms of gene regulation at the translational and transcriptional levels, using the regulation of amino acid–biosynthetic genes in budding yeast as a model system. Transcription of these and many other genes is coordinately induced by the transcriptional activator Gcn4 in response to starvation of any amino acid. Expression of GCN4 is coupled to amino acid levels by a conserved translational control mechanism involving upstream open reading frames (uORFs) in GCN4 mRNA. Ribosomes translate the 5′-most uORF (uORF1) and, under non-starvation conditions, reinitiate translation at uORFs 2, 3, or 4 and then dissociate from the mRNA, keeping GCN4 translation repressed. In starvation conditions, the reinitiating ribosomes bypass uORFs 2–4 and reinitiate at GCN4 instead, owing to decreased availability of the ternary complex (TC)—comprised of initiation factor 2 (eIF2), GTP, and initiator Met-tRNAi—which binds to the small (40S) ribosomal subunit to assemble a 43S preinitiation complex (PIC). TC abundance is reduced in starved cells by phosphorylation of the alpha subunit of eIF2 (eIF2a) by Gcn2, a protein kinase conserved in all eukaryotes, converting eIF2 from substrate to inhibitor of its guanine nucleotide exchange factor (GEF) eIF2B. Hence, GCN4 translation is an in vivo indicator of impaired TC loading on 40S subunits. We previously exploited this fact to isolate mutations in subunits of eIF2B that constitutively derepress GCN4 (Gcd− phenotype) by lowering TC assembly in the absence of eIF2 phosphorylation. More recently, we used the Gcd− selection to identify domains/residues in eIF1, eIF1A, and eIF3, and also residues of 18S rRNA located near the “P” decoding site of the 40S subunit, that participate in rapid TC recruitment in vivo. In collaboration with Jon Lorsch's group, we demonstrated that segments/residues in eIF1, eIF1A, and 18S rRNA, which are implicated genetically in TC recruitment, also stimulate this reaction in a fully reconstituted in vitro system.
We also investigate the roles of various eIFs and the 40S subunit in scanning the mRNA 5′ untranslated region and accurately identifying the AUG initiation codon. The studies exploit a genetic selection for mutations that elevate initiation at near-cognate UUG start codons (Sui− phenotype) or suppress this aberrant initiation event (Ssu− phenotype). In this way, we provided the first evidence that eIF1A and the c-subunit of eIF3 make critical contributions to accurate AUG recognition, and we localized these functions to the unstructured N- and C-tails of eIF1A and the N-terminal domain of eIF3c. In collaboration with Lorsch's group, we also provided strong evidence that dissociation of eIF1 from the PIC is a critical step in AUG recognition, indicating that this factor serves as a "gate-keeper" necessary to suppress initiation at non-AUG triplets. Biochemical analysis revealed that eIF1 stabilizes an "open" conformation of the 40S subunit conducive to scanning and loading of the TC in a metastable state, which enables inspection of successive triplets as they enter the P site for complementarity with the anticodon of Met-tRNAi. However, eIF1 must dissociate from the PIC for AUG selection, as the absence of eIF1 favors the closed, scanning-arrested conformation of the 40S subunit to which the TC is more tightly bound. Our recent analysis of eIF1A provided strong evidence that 10–amino acid repeats in the C-terminal tail (dubbed "scanning enhancers", or SE elements) function together with eIF1 to stabilize the open complex and promote the first step of TC loading, but then must be ejected from the P site to achieve rearrangement to the closed complex, with TC more stably bound and Met-tRNAi base-paired with AUG. Furthermore, we identified scanning-inhibitory (SI) elements in the N-terminal tail and helical domain of eIF1A that oppose the SE elements and promote rearrangement to the closed complex necessary for AUG recognition.
In the arena of transcriptional control, we previously identified multiple co-activators required for gene activation by Gcn4 and defined the molecular program for recruitment of nucleosome-remodeling enzymes and adaptor proteins that remove repressive chromatin structure and recruit TATA–binding protein and other general transcription factors, along with RNA polymerase II (Pol II), to Gcn4 target promoters. We also demonstrated the co-transcriptional recruitment of the histone acetyltransferase (HAT) complexes SAGA and NuA4 to transcribed coding sequences and obtained evidence that they cooperate to promote histone eviction from transcribed coding sequences and to stimulate the rate of Pol II elongation in vivo. We further described a two-stage recruitment mechanism for NuA4, involving the serine-5–phosphorylated C-terminal domain (CTD) of Pol II and methylated histones, and more recently extended the mechanism to include the histone deacetylase (HDA) complex Rpd3C(S). In addition, we demonstrated that the HDAs Rpd3, Hos2, and Hda1 have overlapping functions in deacetylating coding-sequence nucleosomes and observed a strong correlation between increased acetylation and lower histone occupancy in various HDA mutants. The results support the notion that histone acetylation is a key determinant of nucleosome disassembly in transcribed coding sequences.
More recently, we elucidated the roles Pol II CTD kinases Kin28 and Bur1 play in recruitment of the Paf1 complex (Paf1C), an evolutionarily conserved transcription elongation factor for Pol II. Previous work in our lab and of others revealed that Paf1C recruitment is stimulated by the elongation factor DSIF (comprising Spt4 and Spt5) and the CDKs Cdk7/Kin28 and Bur1, which both phosphorylate the Pol II CTD. We also showed that Ser5P CTD phosphorylation by Kin28 enhances recruitment of Bur1 near promoter regions via the pCTD–interaction domain (pCID) in the C-terminus of Bur1, and that Bur1 contributes to serine-2 phosphorylation of the CTD (Ser2P) at the 5′-ends of genes. Accordingly, Kin28 and Bur1 are predicted to collaborate in producing Ser2-/Ser5-diphosphorylated CTD repeats in promoter-proximal Pol II molecules. Our recent findings indicated that Kin28 enhances Paf1C recruitment in two ways, by promoting Bur1 recruitment via the pCTD, with attendant phosphorylation of the Spt5 CTRs by Bur1, and by collaborating with Bur1 to generate Ser2-/Ser5-diphosphorylated CTD repeats that recruit Paf1C via pCIDs in Paf1C subunits capable of recognizing these (and other) diphosphorylated CTD repeats. Interestingly, the pCIDs in certain Paf1C subunits also interact specifically with the phosphorylated Spt5 CTRs. Hence, the Pol II pCTD repeats and Spt5 pCTRs constitute distinct phosphorylated scaffolds in the Pol II elongation complex that cooperate to ensure high-level Paf1C recruitment by Pol II.
β-hairpin loop of eIF1 mediates 40S ribosome binding to regulate Met-tRNAi recruitment and accuracy of AUG selection in vivo.
Recognition of the start codon is thought to require dissociation of eIF1 from the 40S subunit, enabling Pi release by the TC, rearrangement of the 40S subunit to a closed conformation incompatible with scanning, and stable binding of Met-tRNAi to the P site. Supporting this model, we showed that substituting basic amino acids in helix α1 or β-hairpin loop-1, which contact 18S rRNA in the Tetrahymena 40S∙eIF1 crystal structure, impairs eIF1 binding to 40S∙eIF1A complexes in vitro and increases initiation at UUG codons (Sui− phenotype) in a manner suppressed by overexpressing the mutant proteins or an eIF1A mutation (17-21) known to impede eIF1 dissociation in vitro. The eIF1 Sui− mutations also derepress translation of GCN4 mRNA, indicating impaired TC loading, and this Gcd− phenotype is likewise suppressed by eIF1 overexpression or the 17-21 mutation. Thus, direct contacts of eIF1 with 18S rRNA seen in the Tetrahymena 40S∙eIF1 complex are crucial in yeast to stabilize the open conformation of the 40S subunit, required for rapid TC loading and ribosomal scanning, and impede rearrangement to the closed complex at non-AUG codons.
Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex.
Using fluorescence resonance energy transfer (FRET) between fluorophores on the C-terminus of eIF1A and on eIF5 in reconstituted yeast PICs, we found that the C-terminal tail (CTT) of eIF1A moves closer to the N-terminal domain (NTD) of eIF5 in response to start codon recognition in a manner controlled by the rate of eIF1 dissociation from the PIC and dependent on the SE elements in the eIF1A CTT. Remarkably, mutations in the SE elements uncouple eIF1 release from Pi release, dramatically impairing both Pi release and movement of the eIF1A CTT, while minimally affecting eIF1 release. The findings demonstrate that eIF1 dissociation is not sufficient for Pi release and that movement of the eIF1A CTT towards the eIF5 NTD is additionally required for this key step in start codon recognition. We propose that eIF1 release determines the timing of these events by setting the rate of accommodation of tRNAi into the P site, which in turn triggers movement of the eIF1A CTT towards the eIF5 NTD.
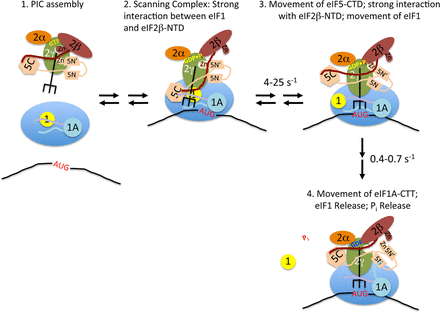
Figure 1. Model for the events taking place within the PIC upon start codon recognition
Stage 1: TC·eIF5 complex binds to the 40S subunit; eIF1 occupies a site on the platform of the 40S adjacent to the P site, and the body of eIF1A binds in the A site, with its NTT (purple) and CTT (light blue) binding in the P site. Stage 2: The scanning PIC is in an open conformation with the tRNAi in the Poutstate; eIF1 binding is stabilized by a strong interaction with the NTT of eIF2β. Stage 3: Entry of the start codon into the P site allows formation of the codon-anticodon helix between the mRNA and tRNAi, which drives the tRNA into the Pin state, which displaces eIF1 to a second, weaker binding site on the 40S subunit, breaking its interaction with the eIF2β NTT, which in turn binds strongly to the eIF5 CTD; movements of the tRNA and/or eIF5 CTD result in changes in the orientation of the eIF5 NTD. Stage 4: eIF1 dissociates from the complex, which, along with accommodation of tRNAi into the P site, causes the CTT of eIF1A to move and interact with the eIF5 NTD; the interaction triggers Pi release from eIF2, possibly by moving the unstructured NTT of eIF5 (not shown for clarity); the resulting complex is in a closed, scanning-arrested state. Reproduced from Nanda et al. (2).
Exome sequencing identifies recurrent somatic mutations in the eIF1A structural gene (EIF1AX) in uveal melanoma with disomy 3.
Uveal melanoma (UM) is the most frequent malignant tumor of the eye, and exome sequencing identified somatic mutations in EIF1AX as prevalent mutations found in UM associated with chromosome 3 disomy. The EIF1AX mutations produce amino acid substitutions or short deletions in the conserved unstructured N-terminal tail (NTT) of eIF1A. Given that we previously showed that NTT substitutions in yeast eIF1A reduce PIC assembly, the EIF1AX mutations could diminish general translation and might also induce transcription factors (e.g., ATF4) whose mRNA translation is inversely coupled to TC concentration by the same mechanism governing translation of yeast GCN4. We also implicated the yeast eIF1A NTT in initiation accuracy, as NTT substitutions suppress initiation at near-cognate triplets (Ssu− phenotype), and cause the PIC to bypass the first AUG encountered while scanning from the 5′ end of the mRNA. Hence, the EIF1AX mutations might suppress recognition of near-cognate initiation sites—which appear to be more prevalent than previously suspected—or of 5′-proximal AUG codons to alter the relative utilization of different start codons for tumor-promoting or tumor-suppressing genes.
Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains.
The factor eIF4B stimulates recruitment of mRNA to the 43S PIC. The mammalian factor stimulates eIF4A helicase activity, but this function is lacking in yeast. The yeast (y)eIF4B consists of: an NTD predicted to be unstructured in solution; an RNA–recognition motif (RRM); and an unusual domain comprising seven imperfect repeats of 26 amino acids; and previous studies implicated the RRM and its RNA binding activity in promoting translation initiation. By analyzing the effects of deletions and mutations of yeIF4B’s domains on PIC attachment to mRNA in vitro and translation initiation in vivo, we found that the 7-repeats domain is critical for productive interaction with the PIC and other components of the initiation machinery, particularly eIF4A, in promoting PIC attachment to mRNA. The NTD also plays a role in accelerating mRNA binding to the PIC but, surprisingly, the RRM and its associated ssRNA binding activity are dispensable in vitro and in vivo. We also found that yeIF4B binds to ribosomal protein Rps20 in the head of the 40S subunit and induces structural changes in the ribosome's mRNA entry channel, suggesting that yeIF4B modulates the 40S head conformation to promote a receptive state of the mRNA entry channel.
Yeast eukaryotic initiation factor 4B (eIF4B) enhances complex assembly between eIF4A and eIF4G in vivo.
Association of eIF4A with eIF4G within eIF4F activates eIF4A helicase activity, as the eIF4G HEAT domains juxtapose the eIF4A RecA-like domains in a conformation poised for catalysis. We identified Ts− mutations in the HEAT domains of yeast eIF4G1 and eIF4G2 that are suppressed by overexpressing either yeIF4B or eIF4A, whereas others are suppressed only by eIF4A overexpression. Importantly, suppression of HEAT domain substitutions by yeIF4B overexpression was correlated with the restoration of native eIF4A∙eIF4G complexes in vivo, and the rescue of specific mutant eIF4A∙eIF4G complexes by yeIF4B was reconstituted in vitro. Association of eIF4A with wild-type (WT) eIF4G in vivo also was enhanced by yeIF4B overexpression and was impaired in cells lacking yeIF4B. Furthermore, we detected native complexes containing eIF4G and yeIF4B but lacking eIF4A. These and other findings lead us to propose that yeIF4B acts in vivo to promote eIF4F assembly by enhancing a conformation of the HEAT domain of yeast eIF4G conducive for stable binding to eIF4A, with attendant recruitment of eIF4A to the mRNA 5′ end to promote 43S PIC attachment.
Roles of vacuolar protein sorting (Vps) factors in regulating transcription
Previously, we reported that disruption of vesicular protein transport at the late endosome/MVB that occur in a subset of vps mutants impedes the ability of nucleus-localized and UAS–bound Gcn4 to stimulate PIC assembly and activate transcription. Recent work by our collaborator Vytas Bankaitis' group led to the model that sterol limitation, or defects in vesicular protein trafficking, provoke a block in Gcn4 function that is triggered by accumulation of sphingolipids [e.g., phytosphingosine (PHS)], in a manner dependent on the sterol-binding protein Kes1 and the Mediator-associated CDK Srb10. Our contribution to this work was to demonstrate by chromatin IP analysis that PHS treatment reduces PIC assembly and Pol II occupancy at the Gcn4 target gene ARG1 to an extent that exceeds the decline in UAS occupancy by Gcn4 at this gene, thus suggesting a transcriptional defect of the type we uncovered previously in certain vps mutants. Bankaitis et al. propose that Kes1 provides a sterol-regulated brake on vesicular protein transport and that increased Kes1 binding to endomembranes at low sterol levels evokes increased cellular levels of sphingolipids (SL) as the result of aberrant expansion of the endosome. The resulting increase in SL levels provokes an Srb10–dependent impairment of Gcn4 activation function, blocking the GAAC response to amino acid starvation in sterol-deprived cells.
Recently, we discovered unexpectedly that a number of Vps factors are associated with the coding sequences of Gcn4 and Gal4 target genes specifically under conditions of transcriptional activation and in a manner dependent on PIC assembly at the associated promoter regions. Consistent with this finding, we uncovered defects in transcriptional elongation in a subset of vps mutants, most notably those lacking the PI(3)P kinase Vps34 and its associated protein kinase Vps15. The phenotypes include reduced mRNA production from long or G+C–rich coding sequences (CDS) without affecting the associated GAL1 promoter activity and a reduced rate of RNA polymerase II (Pol II) progression through lacZ CDS in vivo. Consistent with reported genetic interactions with mutations affecting the histone acetyltransferase complex NuA4, vps15Δ and vps34Δ mutations reduce NuA4 occupancy in certain transcribed CDS. vps15Δ and vps34Δ mutants also exhibit impaired localization of the induced GAL1 gene to the nuclear periphery. We also observed co-localization of a fraction of Vps15-GFP and Vps34-GFP with nuclear pores at nucleus-vacuole (NV) junctions in living cells. The findings suggest that Vps factors enhance the efficiency of transcription elongation in a manner involving their physical proximity to nuclear pores and transcribed chromatin.
Additional Funding
- International Human Frontiers of Science Program Grant RGP0028
Publications
- Martin-Marcos P, Nanda J, Luna RE, Wagner G, Lorsch JR, Hinnebusch AG. β-hairpin loop of eIF1 mediates 40S ribosome binding to regulate initiator tRNAMet recruitment and accuracy of AUG selection in vivo. J Biol Chem 2013;288:27546-27562.
- Nanda JS, Saini AK, Munoz AM, Hinnebusch AG, Lorsch JR. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal pre-initiation complex. J Biol Chem 2013;288:5316-5329.
- Walker SE, Zhou F, Mitchell SF, Larson VS, Valasek L, Hinnebusch AG, Lorsch JR. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA 2013;19:191-207.
- Park EH, Walker SE, Zhou F, Lee JM, Rajagopal V, Lorsch JR, Hinnebusch AG. Yeast eukaryotic initiation factor 4B (eIF4B) enhances complex assembly between eIF4A and eIF4G in vivo. J Biol Chem 2013;288:2340-2354.
- Gaur NA, Hasek J, Brickner DG, Qiu H, Zhang F, Wong CM, Malcova I, Vasicova P, Brickner JH, Hinnebusch AG. Vps factors are required for efficient transcription elongation in budding yeast. Genetics 2013;193:829-851.
Collaborators
- Katsura Asano, PhD, Kansas State University, Manhattan, Kansas, KS
- Vytas Bankaitis, PhD, University of North Carolina School of Medicine, Chapel Hill, NC
- Jason Brickner, PhD, Northwestern University, Evanston, IL
- Juri Hasek, PhD, Institute of Microbiology, Academy of Sciences of the Czech Republic, Prague
- Jon Lorsch, PhD, The Johns Hopkins University School of Medicine, Baltimore, MD
- Gerhard Wagner, PhD, Harvard Medical School, Boston, MA
- Michael Zeschnigk, PhD, Universität Duisburg-Essen, Essen, Germany
Contact
For more information, email hinnebua@mail.nih.gov or visit sncge.nichd.nih.gov.

