Gene Regulation and Cell Biology in the Lyme Disease Pathogen
- Philip P. Adams,
PhD, Head, Group on Gene Regulation in Bacterial Pathogens - Erika Smith, PhD, Postdoctoral Intramural Research Training Award Fellow
- Ryan Fishman, BS, Postbaccalaureate Intramural Research Training Award Fellow
- Robert (Max) Freedman, BS, Postbaccalaureate Intramural Research Training Award Fellow
- Julia Silberman, BS, Postbaccalaureate Intramural Research Training Award Fellow

The goal of this research is to characterize gene regulation and principles of cell biology in the spirochete Borrelia burgdorferi. We use genetics, RNA sequencing, microscopy, and tick-mouse infection models to identify genes for mechanistic study.
The spirochete Borrelia burgdorferi is the causative agent of Lyme disease, an emerging infectious disease and the foremost vector-borne bacterial infection in the world. Given that B. burgdorferi inhabits tick and mammalian hosts, environments with very different temperatures, immune responses, and sources of metabolites, the bacterium must harbor robust gene-regulatory mechanisms in order to survive. Yet, fundamental aspects of gene expression and the basic principles of cell biology have not been studied extensively in B. burgdorferi.
To characterize bacterial regulatory networks, we recognized the importance of mapping RNA boundaries (their 5′ and 3′ ends). Precise identification of RNA ends is critical for gene annotation, the discovery of novel transcripts, and mechanistic characterization of genes. Several RNA-Seq approaches have been developed to globally determine RNA boundaries: 5′RNA-Seq identifies and distinguishes transcription starts sites (TSSs) from 5′ processed ends; total RNA-Seq sequences genes in their entirety; and 3′RNA-Seq captures termination events and identifies RNA 3′ ends. We performed 5′-, total and 3′RNA-Seq on RNA isolated from B. burgdorferi grown in culture [Reference 1]. This identified complex gene arrangements and operons, untranslated regions, and small RNAs. Our findings uncovered an abundance of potential RNA regulators for future study in B. burgdorferi.
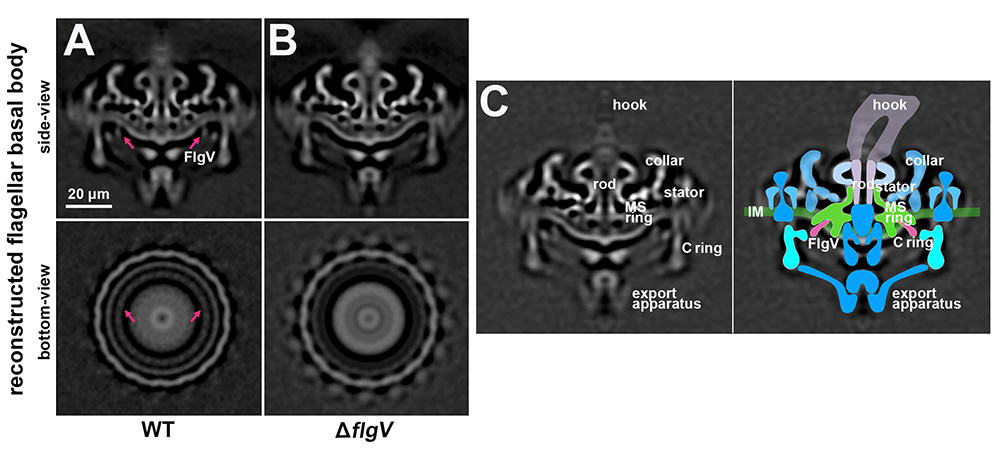
In a separate study, we sought to better understand B. burgdorferi motility [Reference 2]. Flagella propel pathogens through their environments, yet are expensive to synthesize and are immunogenic. Thus, complex hierarchical regulatory networks control flagellar gene expression. Spirochetes are highly motile bacteria, but peculiarly, the archetypal flagellar regulator σ28 is absent from B. burgdorferi. We showed that the gene bb0268 (flgV) is a structural flagellar component that modulates B. burgdorferi flagellar assembly. The flgV gene is broadly conserved in the flagellar superoperon alongside σ28 in many Spirochaetae, Firmicutes and other phyla, with distant homologs in Epsilonproteobacteria. We found that B. burgdorferi FlgV is localized within flagellar basal bodies (Figure 1), and strains lacking flgV produce fewer and shorter flagellar filaments and are defective in cell division and motility. During the enzootic cycle, flgV–deficient B. burgdorferi survive and replicate in Ixodes ticks, but are attenuated for infection and dissemination in mice. Our work defines infection timepoints when spirochete motility is most crucial, and implicates FlgV as a broadly distributed structural flagellar component that modulates flagellar assembly.
Ongoing work in our lab focuses on the characterization and physiological roles of other B. burgdorferi genes, particularly regulatory RNAs and other regulators of motility/cell division.
Figure 1. FlgV is localized to the B. burgforferi flagellar basal body.
Reconstructed cryo-ET side-view and bottom-view cross section of flagellar basal body from (A) wild-type (WT) and (B) ΔflgV B. burgdorferi. (C) Reconstructed cryo-ET flagellar basal body (left) with overlaid cartoon model of the flagellar basal body (right). The inner membrane (IM, green), export apparatus (blue), C ring (light blue), MS ring (green), rod (lavender), hook (light purple), and FlgV (dark pink) are labeled.
Figure 1. FlgV is localized to the B. burgforferi flagellar basal body.
Reconstructed cryo-ET side-view and bottom-view cross section of flagellar basal body from (A) wild-type (WT) and (B) ΔflgV B. burgdorferi. (C) Reconstructed cryo-ET flagellar basal body (left) with overlaid cartoon model of the flagellar basal body (right). The inner membrane (IM, green), export apparatus (blue), C ring (light blue), MS ring (green), rod (lavender), hook (light purple), and FlgV (dark pink) are labeled.
Publications
- Extensive diversity in RNA termination and regulation revealed by transcriptome mapping for the Lyme pathogen Borrelia burgdorferi. Nat Commun 2023 14(1):3931
- Broadly conserved FlgV controls flagellar assembly and Borrelia burgdorferi dissemination in mice. Nat Commun 2024 Preprint
Collaborators
- Ryan Dale, PhD, Computer Support Services Core, NICHD, Bethesda, MD
- Aravind Iyer, PhD, National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD
- Mollie Jewett, PhD, University of Central Florida, Orlando, FL
- Jun Liu, PhD, Yale School of Medicine, New Haven, CT
- Gisela Storz, PhD, Section on Environmental Gene Regulation, NICHD, Bethesda, MD
Contact
For more information, email philip.adams@nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/adams.