The Role of Nuclear Pore Remodeling in Spermatogenesis and Tissue Function
- Mary Dasso,
PhD, Head, Section on Cell Cycle Regulation - Vasilisa Aksenova, PhD, Staff Scientist
- Alexei Arnaoutov, PhD, Staff Scientist
- Maia Ouspenskaia, DVM, Biologist
- Abik Nandi, PhD, Visiting Postdoctoral Fellow
- Sanjana Sundararajan, PhD, Visiting Postdoctoral Fellow
- Reham Aljumaah, BA, Predoctoral Intramural Research Training Award Fellow
- Daniel Cai, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Fatimatou Diouf, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Garrett Thesing, BA, Postbaccalaureate Intramural Research Training Award Fellow

In humans and other eukaryotes, chromosomes reside within the nucleus, a structure surrounded by the double membranes of the nuclear envelope (NE). This organization has profound consequences for gene expression and cell function. The machinery that mediates communication between these compartments thus sits at a crossroads for many pathways of cell regulation and control. Communication between the nucleus and cytoplasm across the NE occurs through conduits called nuclear pore complexes (NPCs), which consist of about 35 proteins called nucleoporins (Figure 1). Beyond nucleocytoplasmic trafficking, nucleoporins are important for chromosome organization, transcriptional control, RNA processing, cell signaling, and cell-cycle control. Moreover, disruption of nucleoporin function can have important clinical implications: nucleoporin genes are frequently mis-regulated in cancers, and nucleoporin mutations cause congenital defects. Interestingly, nucleoporin disruption is linked to phenotypes with a surprisingly high level of tissue specificity, for example, male infertility, pediatric nephrotic syndromes, and neurodegenerative disease.
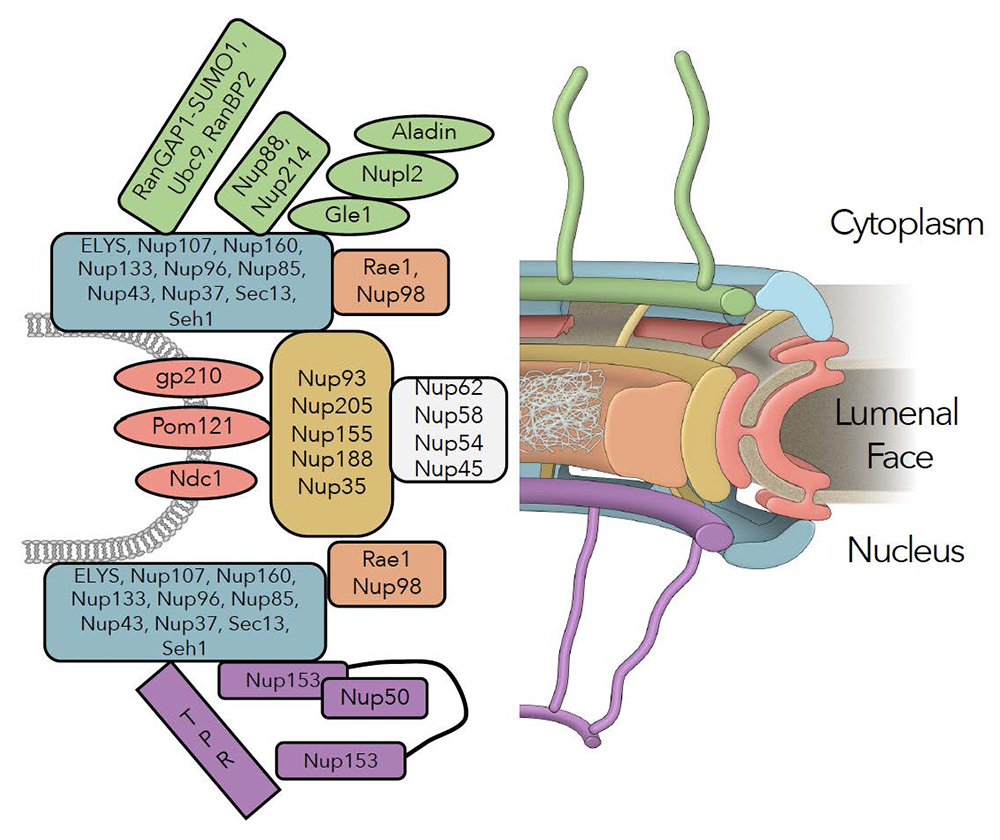
Figure 1. Schematic representation of the NPC
Sub-domains include cytoplasmic fibrils (green), nuclear and cytoplasmic outer rings (blue), the inner ring (tan), Nup98/Rae1 (rust) central channel nucleoporins (white), transmembrane nucleoporins (red), and basket nucleoporins (purple).
Figure 1. Schematic representation of the NPC
Sub-domains include cytoplasmic fibrils (green), nuclear and cytoplasmic outer rings (blue), the inner ring (tan), Nup98/Rae1 (rust) central channel nucleoporins (white), transmembrane nucleoporins (red), and basket nucleoporins (purple).
Our research focuses on understanding the nuclear transport machinery and its impact on processes that are essential for human health. We seek to define the biochemical roles of individual nucleoporins. To assess their role, we adapted auxin-induced degron (AID) strategies for selective and rapid degradation of individual proteins in human tissue-culture cells. This has allowed us to rapidly assess their roles in NPC structure as well as various facets of cellular function. We have also begun to use additional systems, Drosophila and mouse genetics, to monitor how NPCs change during tissue development and to assess clinically relevant nucleoporin phenotypes that are tissues-specific, including nucleoporin roles in the male germline as well as in the glomerulus of the kidney.
The nuclear pore complex (NPC) is a highly modular structure.
Structural models of the NPC depict it as a composite of several sub-domains, which have been named the outer rings, inner ring, cytoplasmic fibrils, and nuclear basket (Figure 1). The distinct roles of individual nucleoporins and their functional interactions remain poorly understood. Moreover, NPCs undergo a disassembly-reassembly cycle during mitotic division, and a lack of tools for acute manipulation of individual nucleoporins had precluded the study of their roles in maintaining structures within pre-existing pores because of complications from disruption of NPC assembly.
We added AID tags and fluorescent moieties by homozygously targeting gene loci encoding Y-complex (components of the NPC) and inner-ring nucleoporins. Auxin addition resulted in a rapid loss of the targeted proteins in each case, without degradation of other nucleoporins. We anticipated that loss of any Y-complex member should result in complete destabilization of the outer rings. While this was true after depletion of NUP96 or NUP107, the loss of other Y-complex members surprisingly left the outer-ring lattice in place. The findings suggest that the outer-ring structure is remarkably resistant to perturbations, once it is fully assembled, and show that its members are not of equivalent importance in sustaining its stability. Furthermore, near-complete loss of the outer ring in NUP96–depleted cells did not cause collapse of the rest of the NPC, as demonstrated by immunostaining, live microscopy, and mass spectrometry. The persistence of inner-ring nucleoporins indicated resilience of the NPC structure. Interestingly, depletion of the inner-ring nucleoporin NUP188 caused an NPC disassembly that was opposite to the profile after NUP96 depletion: inner-ring components were extensively displaced, while the components of the cytosolic fibrils, outer ring, and basket were largely unaffected. Also, there was a global reduction in almost all nucleoporins upon loss of NUP93. Together, our results indicate that the inner and outer rings of the NPC form distinct and independent structures, and that NUP93 serves as an NPC lynchpin essential for both of them.
After depletion of the inner ring or outer rings, we tested whether the residual structures remained functional for the import and export of a model nuclear transport substrate. Remarkably, there were only minimal changes in both nuclear import and export rates upon loss of NUP96 or NUP188. These results indicate that persistent inner-ring or outer-ring structures could still act as conduits for vectoral nuclear trafficking and that these modules can support independent and redundant trafficking routes. The persistence of functional pores lacking a subset of canonical nucleoporins suggests that terminally differentiated cells might retain substantial nuclear trafficking even with divergent NPC composition. Differentiated cells might thus customize function through altered NPC composition, potentially modulating specific trafficking pathways or aspects of NPC activity, such as gene regulation and post-translational protein modifications.
NPC remodeling during spermatogenesis
Spermatogenesis is the remarkable but understudied process for formation of male germ cells, during which the nuclear envelope, chromosomes, and other cellular contents are dramatically remodeled. Importantly, worldwide declines in human sperm counts add urgency to studies of spermatogenesis, both because there is a limited understanding of clinically relevant genes and because a large percentage of human azoospermia cases are classified as idiopathic or arising from de novo changes whose genetic origins remain cryptic. Moreover, there are compelling indications that nucleoporin remodeling is central to male gamete formation. For example, database comparison of human gene expression patterns reveals high expression of nucleoporin-encoding genes in testicular tissues, including nucleoporin variants unique to the testis. Notably, nucleoporin mutations are among the genes implicated in both mammals and flies, where genetic origins of male infertility have been demonstrated. We are currently pursuing studies of spermatogenesis that use flies (Drosophila melanogaster) and mice (Mus musculus) as tractable model organisms with high relevance to human spermatogenesis. These systems will allow us to investigate both the impact of nucleoporin mutations on male fertility, as well as cellular roles of nucleoporins that are unique to the male germline.
Our recent investigations revealed that Drosophila harboring a mutation in nucleoporin dmNup154 (dmNup154R398H, homologous to hsNup155 R391H in humans) exhibit defects in spermatid elongation, likely attributable to disrupted interactions between NPCs and the network of microtubules (MTs) that shape maturing spermatids (Figure 2). Further, our studies suggest a model wherein release of Lamins and basket components of NPCs from the NPC core allows for MT–dependent lateral movement of “chromatin-unanchored” NPC cores along the nuclear envelope, forming a cap-like structure. The structure is reinforced by filamentous actin and actin-binding proteins to create a “dense complex.” Both MT– and actin-dependent forces are essential for subsequent sperm elongation, with the interaction between nucleoporin dmRanBP2 (NPC side) and microtubule motor protein Mst27 (MT side) playing a pivotal role in cap formation and elongation processes. We are currently pursuing two lines of investigation to test this model. First, we are seeking to better characterize the nature of lamina and NPC re-organization during spermatid formation. Second, we will test our model by genetically reconstituting these interactions in other Drosophila tissues in order to test whether they can drive nuclear envelope remodeling outside of the context of the testis. Having established these assays, we further intend to assess the roles of other nucleoporins and nuclear envelope components whose function in the male germline may be critical for fertility (e.g., dmRanGAP, dmRanBP2, dmRae1, Mtor/dmTPR).
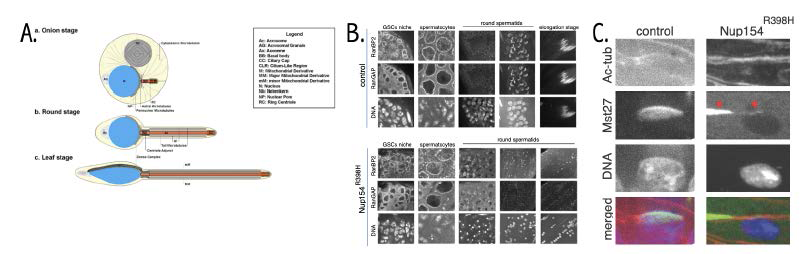
Figure 2. Defective spermatogenesis in Nup154R398H fly testes
A. Early stages of spermatid elongation. Adapted from Riparbelli et al. Cells 2020;9:2684.
B. Localization of RanBP2 and RanGAP in control and Nup154R398H testes. Note correct nuclear pore complex (NPC) staining in both testes during differentiation until the onset of round spermatid stage, where the mutant could not maintain RanBP2 and RanGAP at the nuclear envelope (NE) during initiation of nuclear elongation, and dense complex could not form correctly.
C. Localization of Mst27 and acetylated Tubulin in control and Nup154R398H testes at the stage of initiation of nuclear elongation. Only a single spermatid nucleus is shown. Note that Mst27 (along with RanBP2, not shown) could not be maintained at the nuclear cap (dense complex) and is stripped away from the NE by microtubule (MT) forces (red arrowheads).
Figure 2. Defective spermatogenesis in Nup154R398H fly testes
A. Early stages of spermatid elongation. Adapted from Riparbelli et al. Cells 2020;9:2684.
B. Localization of RanBP2 and RanGAP in control and Nup154R398H testes. Note correct nuclear pore complex (NPC) staining in both testes during differentiation until the onset of round spermatid stage, where the mutant could not maintain RanBP2 and RanGAP at the nuclear envelope (NE) during initiation of nuclear elongation, and dense complex could not form correctly.
C. Localization of Mst27 and acetylated Tubulin in control and Nup154R398H testes at the stage of initiation of nuclear elongation. Only a single spermatid nucleus is shown. Note that Mst27 (along with RanBP2, not shown) could not be maintained at the nuclear cap (dense complex) and is stripped away from the NE by microtubule (MT) forces (red arrowheads).
Our current investigations in mice concern TPR, an evolutionarily conserved nucleoporin that has been implicated in RNA export and accurate mitotic chromosome segregation. While most mammalian tissues have a single predominant isoform of TPR, which we have termed TPR-S, we identified a novel isoform of TPR (TPR-L), which is under positive selection in marsupials and placental mammals (Figure 3). Expression analysis indicated that TPR-L has an alternative start codon and two additional exons and that it is abundantly expressed in human testis in comparison to other tissues. We are working to understand two aspects of TPR-L. First, we are assessing the functional differences between TPR-L and TPR-S through expression of both isoforms in human tissue-culture cells. Second, we generated a TPR-L knockout (KO) mouse lacking TPR-L in all tissues. Knockout males are viable and fertile but show altered seminiferous tubule morphology, observations that suggest that TPR-L loss causes subfertility rather than complete spermatogenic failure. We are currently working to further characterize fertility levels in these animals, particularly as a function of age, as well as analyzing them using transcriptomic analysis and immunostaining for a variety of cell type–specific markers. We hope to expand our mice spermatogenesis studies in the future, based on findings from our work using Drosophila, as well as through analysis of nucleoporins that have been found to be clinically associated with human azoospermia.
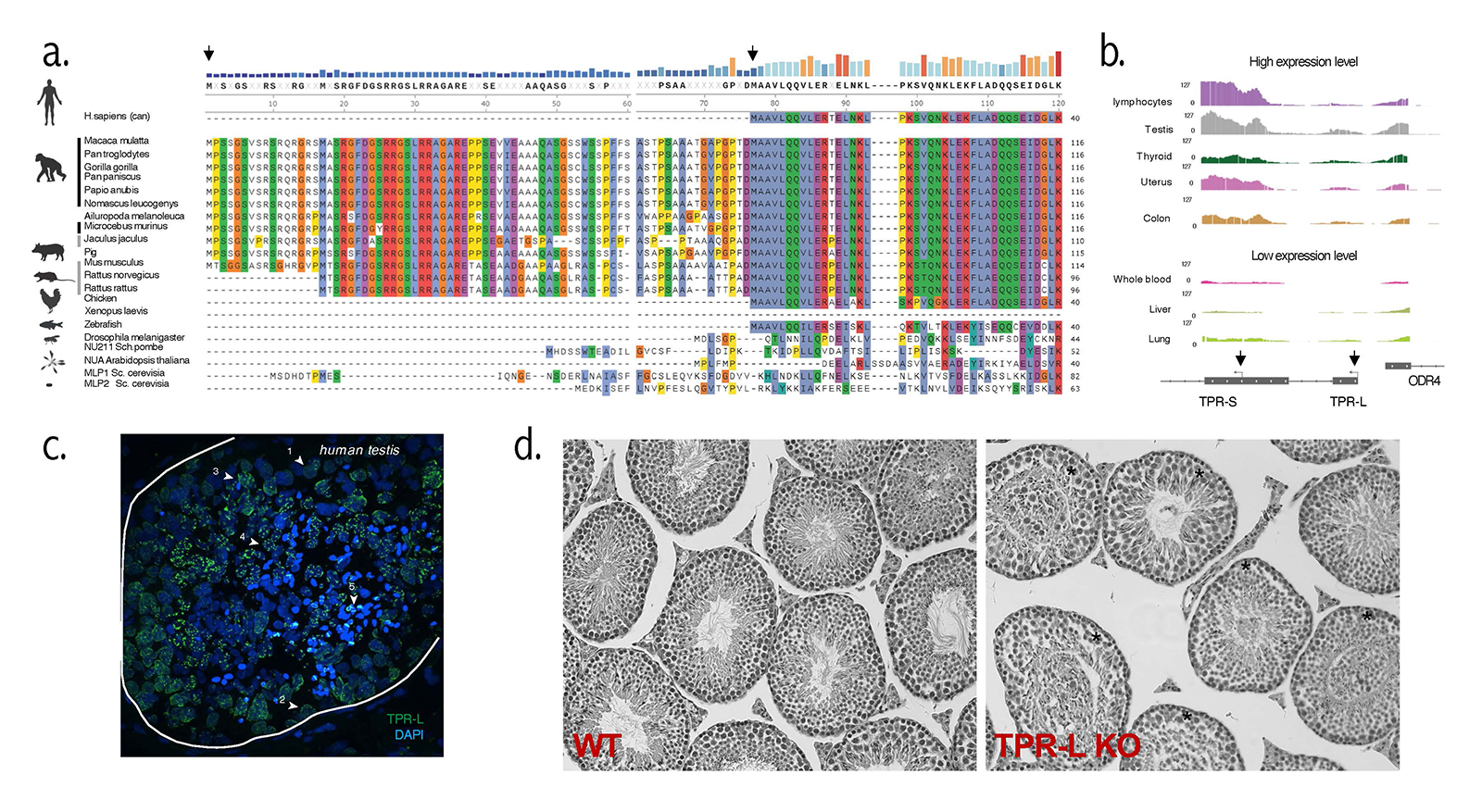
Figure 3. TPR-L in mammalian testes
A. Protein alignment of TPR N-terminus sequence across species.
B. Tissue-specific expression of TPR in GTeX database.
C. Immunostaining of TPR-L (green) within seminiferous tubules. Three somatic and five types of meiotic cells (marked with arrows) express TPR-L.
D. Hematoxylin and eosin staining (decolored) of paraffin-embedded tissue section of wild type (WT) and TPR-L knockout (KO) animals.
Figure 3. TPR-L in mammalian testes
A. Protein alignment of TPR N-terminus sequence across species.
B. Tissue-specific expression of TPR in GTeX database.
C. Immunostaining of TPR-L (green) within seminiferous tubules. Three somatic and five types of meiotic cells (marked with arrows) express TPR-L.
D. Hematoxylin and eosin staining (decolored) of paraffin-embedded tissue section of wild type (WT) and TPR-L knockout (KO) animals.
We anticipate that these studies will contribute toward a better understanding of spermatogenesis and the causes of human azoospermia, an essential step toward the development of better diagnosis and treatment of patients with this condition.
NPC remodeling during podocyte differentiation
Mutations in a variety of nucleoporins have been shown to cause steroid-resistant nephrotic syndrome (SRNS) and a related disorder, the Galloway–Mowat syndrome (GAMOS). SRNS is characterized by proteinuria, increased permeability of the glomerular filtration barrier to proteins that is unresponsive to therapy with steroids. Roughly half the patients with SRNS progress to end-stage renal disease within 15 years, so that it is among the most frequent causes of chronic kidney disease for patients under 30 years old. GAMOS patients similarly show early-onset nephrotic syndrome, in addition to microcephaly and brain anomalies. At a cellular level, these syndromes show disruption of the glomerular structures formed by closely interdigitated podocyte cells that normally serve as filtration barriers in the kidney. Like spermatids, podocytes have an extensively re-organized cytoskeleton, and we have begun to investigate how nucleoporin mutations can disrupt their structure and function. We used mass spectrometry to examine NPC composition in human cell lines that can be induced to differentiate into podocytes or other cell types in vitro. Intriguingly, we observed a distinct composition for the NPCs in podocytes that was not seen in other cell types, with striking changes in the relative levels of nucleoporins that have been genetically implicated in SRNS. We are currently using CRISPR-Cas9 gene editing to introduce mutations found in SRNS patients into our model organisms; we will assess the impact of these mutations on NPC remodeling during podocyte differentiation.
We anticipate that these studies will uncover how NPC structure and function contribute toward podocyte differentiation, how they are disrupted in SRNS patients and why this disruption leads to podocyte loss. This may provide better diagnosis and treatment for SRNS patients and provide insights into other renal diseases in which podocyte function is impacted.
Functional analysis of TREX2 complex subunits and their individual roles in RNA export
A series of evolutionarily conserved complexes are co-transcriptionally recruited to nascent mRNAs, facilitating their processing as well as escorting them to and through the NPC, actions that are functionally linked; a failure to perform any of them during mRNA biogenesis directly impacts both upstream and downstream events. A key player in mRNA maturation is the transcription and export 2 (TREX2) complex, which plays a central role in bridging the transcription and export machinery. The GANP subunit of TREX2 localizes within the nucleus and associates the NPC's nuclear basket, which protrudes from the nucleoplasmic face of the NPC. The TREX2 complex is highly conserved across eukaryotes; The mammalian TREX2 complex consists of GANP (S. cerevisiae homologue is Sac3), PCID2 (Thp1), ENY2 (Sus1), DSS1 (Sem1), and either CETN2 or CETN3 (Cdc31) proteins. In vertebrates, the nuclear basket comprises three nucleoporins, called NUP153, TPR, and NUP50. Our previous studies established that TPR interacts with GANP and that it plays a unique role in the export of TREX2–dependent mRNAs.
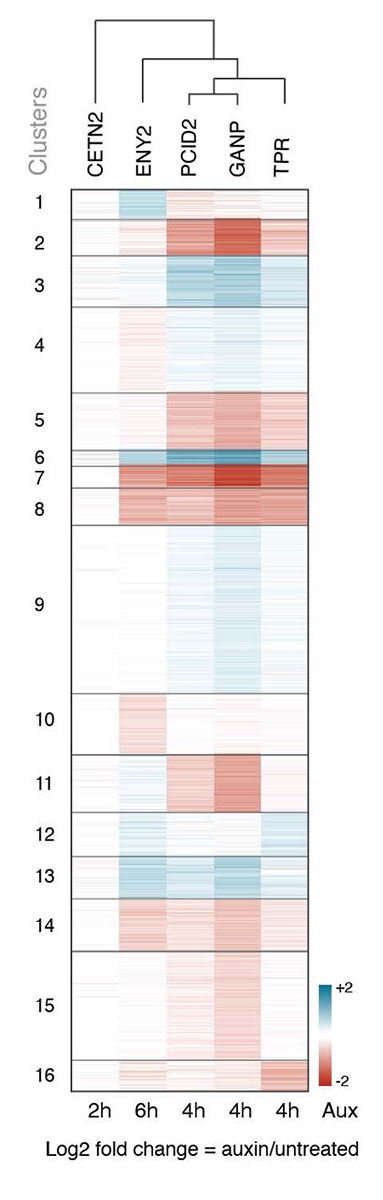
Our current research focuses on the roles of individual TREX2 complex subunits in cell lines in which PCID2, ENY2, CETN2 and CETN3 were tagged with AID-NeonGreen. We found that depletion of different subunits caused distinct transcriptomic changes and impacted RNA export from the nucleus to different extents, suggested that TREX2 subunits do not operate in perfect tandem. We hypothesize that they may form different functional complexes, whose composition and assembly can be determined by the protein partners or RNAs present in a particular cell type. We are currently analyzing the biochemical binding partners of individual TREX2 subunits.
As we learn about the biochemical roles of TREX-2 subunits in tissue culture cells, we will extend these studies toward understanding of how TREX-2 function is remodeled in different tissues and whether such changes contribute toward altered gene expression during differentiation.
Additional Funding
- FY24 Developing Talent Scholarship (Fatimatou Diouf)
- FY24 NICHD Career Development Award for Staff Scientists/Staff Clinicians (Vasilisa Aksenova))
- FY24 NICHD Intramural Research Fellowship (Sanjana Sundararajan)
Publications
- Interphase chromosome conformation is specified by distinct folding programs inherited via mitotic chromosomes or through the cytoplasm. bioRxiv 2024 PMID: 39345587
- Cellular NS1-BP protein interacts with the mRNA export receptor NXF1 to mediate nuclear export of influenza virus M mRNAs. J Biol Chem 2024 300:107871
- Methylated histones on mitotic chromosomes promote Topoisomerase IIα function for high-fidelity chromosome segregation. iScience 2023 26:106743
- Influenza virus mRNAs encode determinants for nuclear export via the cellular TREX-2 complex. Nat Commun 2023 14:2304
- Ectopic expression of meiotic cohesin generates chromosome instability in cancer cell line. Proc Natl Acad Sci USA 2022 119:e2204071119
- Architecture of the cytoplasmic face of the nuclear pore. Science 2022 376:6598
Collaborators
- Yoshiaki Azuma, PhD, University of Kansas, Lawrence, KS
- Job Dekker, PhD, University of Massachusetts Chan Medical School, Worcester, MA
- Beatriz Fontoura, PhD, University of Texas Southwestern Medical Center, Dallas, TX
- André Hoelz, PhD, California Institute of Technology, Pasadena, CA
- Yi Ren, PhD, Center for Structural Biology, Vanderbilt University School of Medicine, Nashville, TN
- Alexander Strunnikov, PhD, Guangzhou Institutes of Biomedicine and Health, Guangzhou, China
Contact
For more information, email dassom@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/dasso.