Transcriptional and Translational Regulatory Mechanisms in Nutrient Control of Gene Expression
- Alan G. Hinnebusch,
PhD, Head, Section on Nutrient Control of Gene Expression - Fan Zhang, MS, Senior Research Assistant
- Darren Fenton, PhD, Postdoctoral Fellow
- Ritu Gupta, PhD, Postdoctoral Fellow
- Rakesh Kumar, PhD, Postdoctoral Fellow
- Priyanka Mittal, PhD, Postdoctoral Fellow
- Poonam Poonia, PhD, Postdoctoral Fellow
- Priyanka Singh, PhD, Postdoctoral Fellow
- Sakshi Singh, PhD, Postdoctoral Fellow
- Vishalini Valabhoju, PhD, Postdoctoral Fellow
- Anil Vijjamarri, PhD, Postdoctoral Fellow
- Brittany Benner, BS, Postbaccalaureate Fellow
- Hana Lee, BS, Postbaccalaureate Fellow

We study molecular mechanisms underlying the assembly, function, and regulation of translation initiation complexes involved in protein synthesis, using yeast as a model system to exploit its powerful combination of genetics and biochemistry. The translation initiation pathway produces an 80S ribosome bound to mRNA with methionyl initiator tRNA (tRNAi) base-paired to the AUG start codon in the ribosomal P site. The tRNAi is recruited to the 40S subunit in a ternary complex (TC) with GTP–bound eIF2 to produce the 43S preinitiation complex (PIC) in a reaction stimulated by eIFs 1, 1A, 3, and 5. The 43S PIC attaches to the 5′ end of mRNA, facilitated by the cap-binding complex eIF4F (comprising eIF4E, eIF4G, and RNA helicase eIF4A) and poly(A)–binding protein bound to the poly(A) tail, and scans the 5′ UTR for an AUG start codon in preferred sequence context. Scanning is promoted by eIFs 1 and 1A, which induce an open conformation of the 40S, and by eIF4F and RNA helicase Ded1, which resolve RNA structures in the 5′ UTR. AUG recognition stabilizes a closed conformation of the PIC, dissociation of eIF1, highly stable P-site binding of tRNAi, and hydrolysis of the GTP bound to eIF2 stimulated by eIF5. Subsequent release of eIF2–GDP from tRNAi allows joining of the 60S subunit to form the 80S initiation complex. eIF2 function is down-regulated by phosphorylation of its alpha subunit by protein kinase Gcn2 in response to amino acid starvation and other stresses that likely evoke stalling of ribosomes engaged in translation elongation.
Our current aims in this research area are to: (1) elucidate the functions of eIF1, eIF5, eIF3, and 40S ribosomal proteins in TC recruitment and start-codon recognition; (2) identify distinct functions of the RNA helicases eIF4A (and its cofactors eIF4G/eIF4B), Ded1, and Dbp1, and of the poly(A)–binding protein (PABP) in mRNA activation, 48S PIC assembly, and scanning in vivo; (3) uncover the mechanisms of translational repression and regulation of mRNA abundance by the repressors Scd6, Pat1, the helicase Dhh1, and the mRNA–decapping enzyme Dcp2, and identify the RNA–binding proteins involved in these functions; (4) elucidate the regulation of Ded1, eIF4G, and Dhh1 functions in response to nutrient limitation or stress; (5) elucidate the roles of the yeast orthologs of eIF2A and eIF2D in eIF2–independent initiation of translation and reinitiation in stress conditions; (6) elucidate the mechanism of activation of the protein kinase Gcn2 in response to amino acid starvation and perturbations of translation elongation; and (7) elucidate the role of eIF4E–binding protein Eap1 in regulating mRNA decay and translation.
We also study fundamental mechanisms of transcriptional control of gene expression by analyzing the coordinate induction of more than 100 amino acid–biosynthetic genes by activator Gcn4 during amino acid limitation. We and others showed previously that Gcn4 recruits multiple cofactor complexes including SAGA, which contains the histone acetyltransferase (HAT) Gcn5, the chromatin remodeling (CR) complexes SWI/SNF and RSC, and the RNA Polymerase II (Pol II) Mediator, which in turn stimulate recruitment of general transcription factors (GTFs), including TATA–binding protein (TBP), and Pol II to promoter DNA to stimulate preinitiation complex (PIC) assembly. Seeking to identify all cofactors that mediate eviction of the -1 and +1 nucleosomes that occlude promoter DNA and impede binding of TBP and PIC assembly, we recently uncovered extensive cooperation among SWI/SNF, RSC, and a third CR, Ino80C, in evicting and repositioning promoter nucleosomes at genes activated by Gcn4 and other highly expressed genes in starved cells. We are currently probing the relative contributions of co-factors SAGA, TFIID, and Mot1 in recruitment of TBP to gene promoters throughout the genome, with a particular focus on the Gcn4 transcriptome in amino acid–starved cells.
Non-canonical initiation factor eIF2A has a minimal role in translation initiation even in cells with reduced eIF2 function.
It has been reported that mammalian eIF2A promotes Met-tRNAi recruitment to the AUG codons of viral mRNAs and enhances initiation at near-cognate codons (NCCs) when TC assembly is impaired by eIF2α phosphorylation. We conducted ribosome profiling on a yeast eIF2AΔ mutant in the presence or absence of eIF2α phosphorylation induced by amino acid starvation, reasoning that mRNAs able to utilize eIF2A might show TE reductions only when eIF2 is impaired. Surprisingly, eIF2AΔ conferred no significant TE reductions for any mRNAs in non-starved cells, and affected only about 30 mRNAs in amino acid–starved cells. We were able to validate this conditional eIF2A dependence by luciferase (LUC) reporter analysis and polysome profiling of native mRNAs for only several genes. We found no evidence that eIF2AΔ alters the translation of mRNAs containing evolutionarily conserved, inhibitory uORFs. Our findings do not support an important role for yeast eIF2A in a back-up mechanism for Met-tRNAi recruitment when eIF2 function is reduced by stress.
Differential requirements for ribosomal P-stalk components in activating yeast protein kinase Gcn2 by stalled ribosomes during stress
A highly conserved response to amino acid starvation involves activation of the protein kinase Gcn2, which phosphorylates eukaryotic initiation factor 2, with attendant inhibition of global protein synthesis and increased translation of the yeast transcriptional activator GCN4. Gcn2 contains a domain related to histidyl-tRNA synthetase (HisRS–like domain), and a C-terminal ribosome-binding domain. Previous work indicated that Gcn2 is activated on translating ribosomes by uncharged tRNAs that accumulate in amino acid–starved cells and pair with the cognate codons in the empty ribosomal A, interacting directly with the HisRS–like domain to stimulate kinase activity. Gcn2 can also be activated by conditions that stall elongating ribosomes without reducing aminoacylation of tRNA, but it was unclear whether distinct molecular mechanisms operate in these two circumstances. We identified three regimes that activate Gcn2 in yeast by starvation-independent (SI) ribosome stalling, which leaves an empty A site on the stalled ribosome: (1) treatment with inhibitor tigecycline, which should stall ribosomes at all codons; (2) deleting the gene encoding tRNAArgUCC, which should stall ribosomes at AGG codons; and (3) depletion of translation-termination factor eRF1, which should stall ribosomes at stop codons. Subsequent genetic analysis demonstrated requirements for the HisRS–like and ribosome-binding domains of Gcn2, positive effectors Gcn1/Gcn20, and the tethering of at least one of two P1/P2 heterodimers of the 60S ribosomal P-stalk complex, for detectable activation by SI–ribosome stalling. Remarkably, no tethered P1/P2 proteins were required for strong Gcn2 activation by starvation for various amino acids, indicating that Gcn2 activation has different requirements for the P-stalk, depending on how ribosomes are stalled. We propose that accumulation of deacylated tRNAs in starved cells functionally substitutes for the P-stalk in binding to the HisRS–like domain for eIF2 kinase activation by ribosomes stalled with A sites.
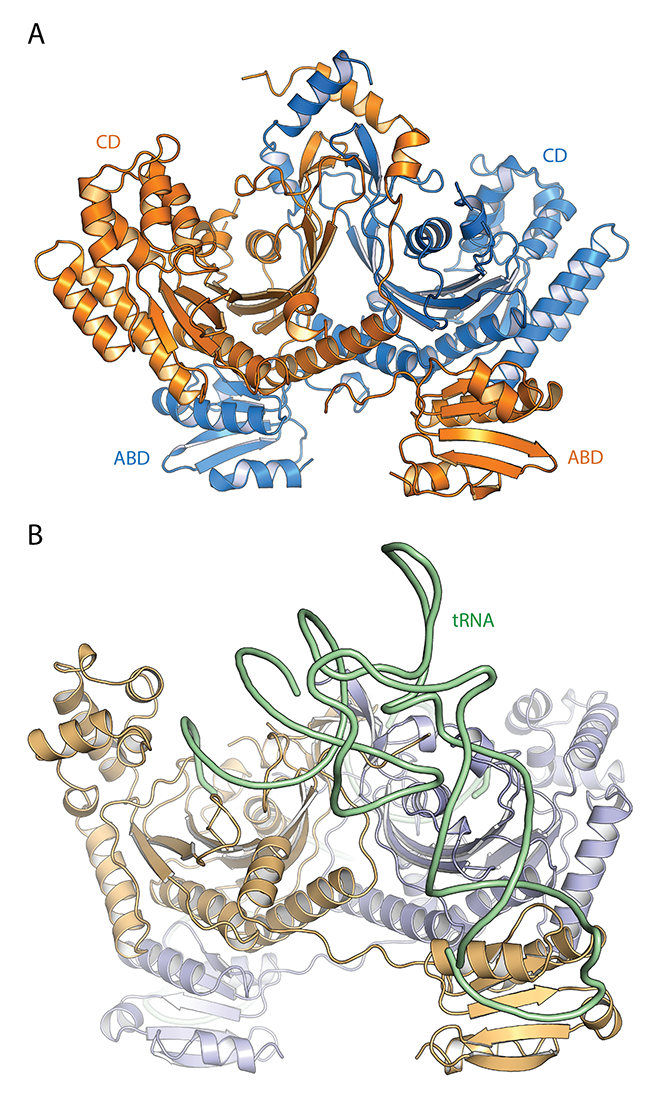
Figure 1. Structural mimicry of the histidyl-tRNA charging enzyme (HisRS) by the HisRS–related sensor of amino acid starvation in Gcn2, the ancestral protein kinase of the Integrated Stress Response
A. Overall crystal structure of the C. thermophilum Gcn2 HisRS–related regulatory domain, containing structural mimics of both the catalytic domain (CD), where the 3′ acceptor end of tRNA is aminoacylated, and the anticodon binding domain (ABD), which binds the anticodon loop of tRNA to ensure its identify for aminoacylation.
B. Co-crystal structure of T. thermophilus authentic HisRS enzyme bound to histidyl tRNA (PDB:4RDX). We showed that substituting conserved ABD residues in the Gcn2 HisRS–related domain, well positioned to contact the tRNA anticodon loop, impairs both Gcn2 function in starved cells and eIF2 phosphorylation by purified Gcn2 in vitro. Mimicry in Gcn2 of two highly conserved structural domains for binding both ends of tRNA, each crucial for Gcn2 function, supports the model that deacylated tRNAs activate Gcn2 in amino acid–starved cells and exemplifies how a metabolic enzyme is repurposed to host new local structures and sequences that confer a novel regulatory function.
Figure 1. Structural mimicry of the histidyl-tRNA charging enzyme (HisRS) by the HisRS–related sensor of amino acid starvation in Gcn2, the ancestral protein kinase of the Integrated Stress Response
A. Overall crystal structure of the C. thermophilum Gcn2 HisRS–related regulatory domain, containing structural mimics of both the catalytic domain (CD), where the 3′ acceptor end of tRNA is aminoacylated, and the anticodon binding domain (ABD), which binds the anticodon loop of tRNA to ensure its identify for aminoacylation.
B. Co-crystal structure of T. thermophilus authentic HisRS enzyme bound to histidyl tRNA (PDB:4RDX). We showed that substituting conserved ABD residues in the Gcn2 HisRS–related domain, well positioned to contact the tRNA anticodon loop, impairs both Gcn2 function in starved cells and eIF2 phosphorylation by purified Gcn2 in vitro. Mimicry in Gcn2 of two highly conserved structural domains for binding both ends of tRNA, each crucial for Gcn2 function, supports the model that deacylated tRNAs activate Gcn2 in amino acid–starved cells and exemplifies how a metabolic enzyme is repurposed to host new local structures and sequences that confer a novel regulatory function.
Structural evidence that uncharged tRNA is an activator of Gcn2
We collaborated with Jinwei Zhang to determine the crystal structure of the HisRS–like domain of Gcn2 from the fungus Chaetomium thermophilum. The structure is dramatically similar to authentic HisRS, including an α-helical insertion domain harboring the catalytic center and a separate anticodon binding domain (ABD) that, in authentic HisRS, bind to the acceptor stem and anticodon loop of tRNAHis, respectively. The structural elements for forming histidyl adenylate and aminoacylation of tRNAHis are lacking, consistent with repurposing of the HisRS–like domain as a sensor of amino acid starvation. This model is supported by the location of numerous substitutions in the conserved catalytic core of the HisRS–like domain that hyperactivate or impair Gcn2 function. We further showed that residues in the ABD predicted by the structure to contact the tRNA anticodon loop or connect the ABD to the catalytic core are also crucial for Gcn2 activation in vivo and high-level kinase activity by purified Gcn2 in vitro. The presence of two conserved structural domains for binding different ends of tRNA in the HisRS–like domain, both essential for kinase activation, supports the model that uncharged tRNA directly activates Gcn2 on stalled ribosomes in amino acid–starved cells.
Yeast-specific roles for Tma20/Tma22 in controlling reinitiation on GCN4 mRNA
Evidence suggests that the mammalian heterodimer MCT-1/DENR promotes reinitiation (REI) following translation of short uORFs whose penultimate codons depend on the heterodimer for releasing the cognate deacylated tRNA from post-termination 40S complexes (postTCs), including uORF1 of ATF4 mRNA, which mediates the same delayed REI mechanism governing GCN4 translation. Substituting GCN4 uORF1 with penultimate codons that vary in dependence on the corresponding translation machinery–associated protein heterodimer Tma20/Tma22 for 40S recycling, we found little evidence that Tma20/Tma22 enhances REI at GCN4 uORF1 variants equipped with Tma–dependent penultimate codons. Making similar substitutions at uORF4, which differs from uORF1 in being permissive for REI, we implicated Tma20/Tma22 in blocking, not stimulating REI, by releasing empty postTCs, in a manner only marginally more pronounced with Tma20/Tma22–dependent penultimate codons. Thus, dissociation of tRNA from postTCs can proceed without Tma20/Tma22 at both uORF1 and uORF4, even at Tma–dependent penultimate codons, and Tma20/Tma22 enhances dissociation of postTCs at uORF4 variants with either class of penultimate codons. The latter function is impeded at uORF1 by its specialized REI–promoting elements. We conclude that Tma20/Tma22 and MCT-1/DENR play distinct roles in the delayed REI mechanism.
Differential requirements for Gcn5 and NuA4 HAT activities in the starvation-induced versus basal transcriptomes
Previously, we showed that elimination of Gcn5, the histone acetyltransferase (HAT) subunit in co-factor SAGA (Spt-Ada-Gcn5 acetyltransferase), did not fully impair nucleosome eviction at many starvation-induced genes, suggesting that Gcn5 might cooperate with other HATs in this process, similar to the functional cooperation we had identified for different CRs (chromatin remodelers). The role of the HAT complex NuA4, responsible for most H4 and H2A acetylation in yeast, was of particular interest, as it was shown to be recruited to the Gcn4 target genes ARG1 and ARG4. We examined the effects of disrupting the NuA4 complex, by eliminating its nonessential scaffold subunit Eaf1, on promoter nucleosome eviction and transcriptional activation at both starvation-induced and constitutively expressed genes. We also examined whether depleting Eaf1 from the nucleus (by anchor-away technology) confers defects in nucleosome eviction or transcription in cells lacking Gcn5 in order to evaluate whether NuA4 and Gcn5 make independent, additive contributions to these processes at particular genes in vivo. Our results revealed that NuA4 acts on par with Gcn5, and functions additively, in evicting and repositioning promoter nucleosomes, and in stimulating transcription, at starvation-induced genes. However, NuA4 is generally more important than Gcn5 in promoter nucleosome eviction, recruitment of the TATA-binding protein (TBP), and transcription at most other genes expressed constitutively in yeast. NuA4 also predominates over Gcn5 in stimulating TBP recruitment and transcription of genes categorized as principally dependent on the cofactor TFIID versus SAGA, except for the highly expressed subset encoding ribosomal proteins (RPs), where Gcn5 contributes strongly to PIC assembly and transcription. We found that both SAGA and NuA4 are recruited to promoter regions of starvation-induced genes in a manner that appears to be controlled by their HAT activities, and thus most likely act directly to promote transcription of these genes. Our findings reveal an intricate interplay between these two HATs in nucleosome eviction, PIC assembly, and transcription that differs between the starvation-induced and basal transcriptomes.
Publications
- Differential requirements for P-stalk components in activating yeast protein kinase Gcn2 by stalled ribosomes during stress. Proc Natl Acad Sci USA 2023 120:e2300521120
- Differential requirements for Gcn5 and NuA4 HAT activities in the starvation-induced versus basal transcriptomes. Nucleic Acids Res 2023 51:3696–3721
- Yeast eIF2A has a minimal role in translation initiation and uORF-mediated translational control in vivo. eLife 2024 12:RP92916
- Differential effects of 40S ribosome recycling factors on reinitiation at regulatory uORFs in GCN4 mRNA are not dictated by their roles in bulk 40S recycling. Commun Biol 2024 7:1083
- Gcn2 structurally mimics and functionally repurposes the HisRS enzyme for the integrated stress response. Proc Natl Acad Sci USA 2024 121:e2409628121
Collaborators
- Paul Cullen, PhD, Department of Biological Sciences, State University of Buffalo, Buffalo, NY.
- Miriam L. Greenberg, PhD, Department of Biological Sciences, Wayne State University, Detroit, MI
- Zhenguo Lin, PhD, Department of Biology, Saint Louis University, St. Louis, MO
- Jose L. Llácer, PhD, Instituto de Biomedicina de Valencia (IBV-CSIC), Valencia, Spain
- Jon Lorsch, PhD, Laboratory on the Mechanism and Regulation of Protein Synthesis, NICHD, Bethesda, MD
- Neelam Sen, PhD, Jawaharlal Nehru University, New Delhi, India
- Jinwei Zhang, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
Contact
For more information, email hinnebua@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/hinnebusch.