Molecular Genetics of an Imprinted Gene Cluster on Mouse Distal Chromosome 7
- Karl Pfeifer,
PhD, Head, Section on Epigenetics - Claudia Gebert, PhD, Biologist
- Beenish Rahat, PhD, Postdoctoral Fellow
- Hyungchul Lee, PhD, Visiting Fellow
- Abucar Mohamed, BS, Postbaccalaureate Fellow

Our lab is interested in epigenetic mechanisms that regulate gene expression in order to understand how the epigenome is established and then modified during development and how disruptions in the epigenome can lead to developmental disorders and disease. A simple definition of the epigenome is that it is the total array of heritable changes in chromosome structures that influence gene function. Known epigenetic mechanisms include DNA modifications (such as cytosine methylation), altering histone localization and chemical structures, and DNA looping. Our approach is to build and then characterize mouse models for human disorders in which the fundamental problem is disruption of the normal epigenome. By understanding disease progression in these models, we hope to learn about mechanisms for regulating the epigenome. But we also hope that clarifying the exact molecular defects in these specific diseases will identify new therapeutic targets. We are currently investigating two disease models. First, we are continuing our long-term and comprehensive analysis of maternal loss of imprinting (LOI) at the H19/Igf2 locus. As detailed in the next paragraph, maternal LOI is a model for the Beckwith-Wiedeman syndrome, the most common overgrowth and cancer predisposition disorder. Second, we are generating and characterizing new mouse models that disrupt cohesin accumulation on the chromosomes. Altered cohesin patterns are the molecular cause for the genetic disorder called Cornelia-de-Lange syndrome.
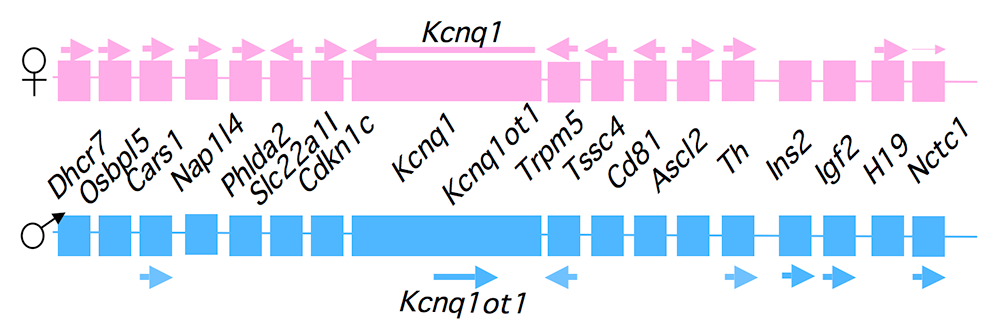
Genomic imprinting is an unusual form of gene regulation by which an allele’s parental origin restricts allele expression. For example, almost all expression of the noncoding RNA tumor-suppressor gene H19 is from the maternal chromosome. In contrast, expression of the neighboring insulin-like growth factor 2 gene (Igf2) is from the paternal chromosome. Imprinting represents the cleanest possible example of epigenetic regulation of gene expression, and therefore understanding imprinting is an excellent means to understand epigenetics. Imprinted genes are not randomly scattered throughout the chromosome but rather are localized in discrete clusters, where monoallelic expression is regulated by a common cis-acting DNA–regulatory element called the Imprinting Control Region (ICR). We study a cluster of imprinted genes on the distal end of mouse chromosome 7 (Figure 1). The syntenic region in humans (11p15.5) is highly conserved in gene organization and expression patterns. Imprinting of H19 and Igf2 is regulated by the H19 ICR, which is located just upstream of the H19 promoter. We showed that the molecular function of the H19 ICR is to organize the region into alternative 3D structures. In humans, epigenetic mutations that disrupt H19 ICR function result in loss of monoallelic expression. Mutations in the paternal H19 ICR lead to loss of Igf2 expression and biallelic (2X) H19 expression, and are associated with the Silver-Russell syndrome (intrauterine growth restriction). Mutations in the maternal H19 ICR lead to loss of H19 but biallelic (2x) Igf2 expression, and are associated with the Beckwith-Wiedemann syndrome and several pediatric cancers. Our lab generated mouse models that phenocopy the human diseases, and our goal is to characterize the molecular defects associated with mis-expression of Igf2/H19 and to understand how these molecular defects lead to disease and cancer.
Alternative long-range interactions between distal regulatory elements establish allele-specific expression at the Igf2/H19 locus.
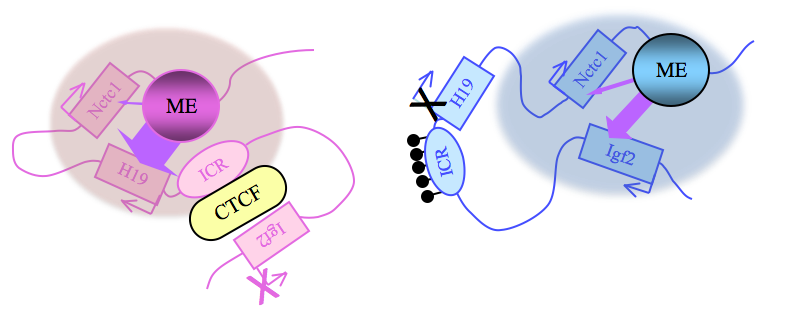
Paternally expressed Igf2 lies about 80 kb upstream of the maternal-specific H19 gene. Using cell-culture systems as well as transgene and knockout experiments in vivo, we identified the enhancer elements responsible for activation of the two genes. The elements are shared and are all located downstream of the H19 gene (Figure 2). Imprinting at the Igf2/H19 locus depends on the 2.4 kb H19 ICR, which lies between the two genes, just upstream of the H19 promoter (Figure 2). On the maternal chromosome, binding of the CTCF protein, a transcriptional repressor, to the H19 ICR establishes a transcriptional insulator that organizes the chromosome into loop structures that bring the H19 promoter into contact with downstream enhancers but exclude the Igf2 promoter from these enhancer interactions. The loops favor H19 expression but block interactions between the maternal Igf2 promoters and the downstream shared enhancers, thus preventing maternal Igf2 expression. Upon paternal inheritance, the cytosine residues within the ICR DNA sequences are methylated, which prevents binding of the CTCF protein, so that a transcriptional insulator is not established. Thus, paternal Igf2 promoters and the shared enhancers interact via DNA loops, and expression of paternal Igf2 is facilitated. Taken together, we find that the fundamental role of the ICR is to organize the chromosomes into alternative 3-D configurations that promote or prevent expression of the Igf2 and H19 genes.
Figure 2. Distinct maternal and paternal chromosomal conformations at the distal 7 locus
Epigenetic modifications on the 2.4 kb ICR generate alternative 3D organizations across a large domain on paternal (blue) and maternal (pink) chromosomes and thereby regulate gene expression. ICR, imprinting control region; ME, muscle enhancer; filled lollipops, CpG methylation covering the paternal ICR.
Figure 2. Distinct maternal and paternal chromosomal conformations at the distal 7 locus
Epigenetic modifications on the 2.4 kb ICR generate alternative 3D organizations across a large domain on paternal (blue) and maternal (pink) chromosomes and thereby regulate gene expression. ICR, imprinting control region; ME, muscle enhancer; filled lollipops, CpG methylation covering the paternal ICR.
The H19 ICR is not only necessary but also sufficient for genomic imprinting. To demonstrate this, we used knock-in experiments to insert the 2.4 kb element at heterologous loci and demonstrated its ability to imprint these regions. Furthermore, analyses of the loci confirmed and extended the transcriptional model described above. Upon maternal inheritance, even ectopic ICR elements remain unmethylated, bind to the CTCF protein, and form transcriptional insulators. Paternally inherited ectopic ICRs become methylated, cannot bind to the CTCF, and therefore promote alternative loop domains distinct from those organized on maternal chromosomes. Our most curious finding was that DNA methylation of ectopic ICRs is not acquired until relatively late in development, after the embryo implants in the uterus. In contrast, at the endogenous locus, ICR methylation occurs during spermatogenesis. The findings thus imply that DNA methylation is not the primary imprinting mark that distinguishes maternally from paternally inherited ICRs.
The Nctc1 gene lies downstream of H19 and encodes a spliced, polyadenylated long noncoding RNA (lncRNA), which is transcribed across the muscle enhancer element (ME in Figure 2); the element is shared by Igf2 and H19. Nctc1 expression depends on this enhancer element. Concordantly, the shared enhancer interacts with the Nctc1 promoter, just as it interacts with the maternal H19 and paternal Igf2 promoters. We showed that all three co-regulated promoters (Igf2, H19, and Nctc1) also physically interact with each other in a manner that depends on their interactions with the shared enhancer. Thus, enhancer interactions with one promoter do not preclude interactions with another promoter. Moreover, we demonstrated that such promoter-promoter interactions are regulatory; they explain the developmentally regulated imprinting of Nctc1 transcription. Taken together, our results demonstrate the importance of long-range enhancer-promoter and promoter-promoter interactions in physically organizing the genome and establishing the gene expression patterns that are crucial for normal mammalian development.
Molecular mechanisms for tissue-specific promoter activation by distal enhancers
Normal mammalian development is absolutely dependent on establishing the appropriate patterns of expression of thousands of developmentally regulated genes. Most often, development-specific expression depends on promoter activation by distal enhancer elements. The Igf2/H19 locus is a highly useful model system for investigating mechanisms of enhancer activation. First, the biological significance of the model is clear, given that expression of these genes is so strictly regulated. Even twofold changes in RNA levels are associated with cancer and developmental disorders. Second, we already know much about the enhancers in this region and have established powerful genetic tools to investigate their function. Igf2 and H19 are co-expressed throughout embryonic development and depend on a series of tissue-specific enhancers that lie between 8 kb and more than 150 kb downstream of the H19 promoter (or between 88 kb and more than 130 kb downstream of the Igf2 promoters). The endodermal and muscle enhancers have been precisely defined, and we generated mouse strains carrying deletions that completely abrogate enhancer function. We also generated insulator insertion mutations that specifically block muscle enhancer activity and used these strains to generate primary myoblast cell lines so that we can combine genetic, molecular, biochemical, and genomic analyses to understand the molecular bases for enhancer functions.
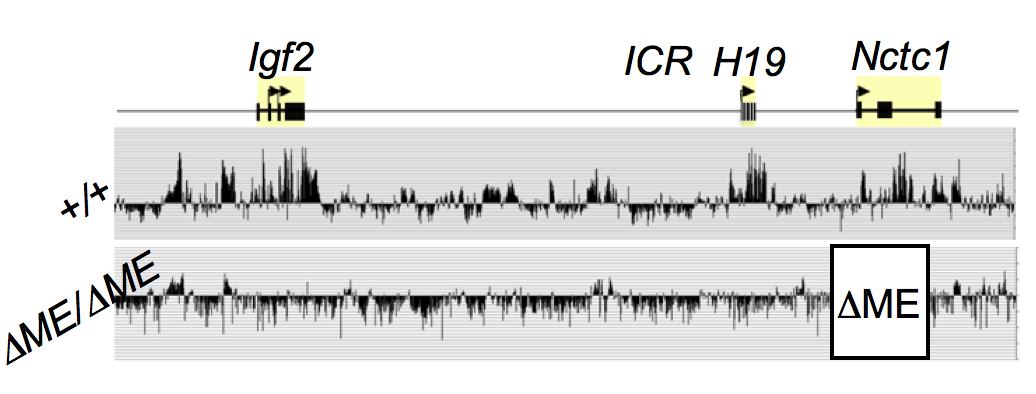
The muscle enhancer (ME) directs RNA polymerase (RNAP) II not only to its cognate promoters (i.e., to the H19 and Igf2 promoters) but also across the entire intergenic region. To demonstrate this, we used ChIP-on-chip to analyze RNAP localization on chromatin prepared from wild-type and enhancer-deletion (DME) cell lines (Figure 3). As expected, RNAP binding to the H19 and Igf2 promoters is entirely enhancer-dependent. Curiously, we also noted enhancer-dependent RNAP localization across the entire locus, including the large intergenic domain between the two genes. Furthermore, RNAP binding is associated with RNA transcription. Thus, the enhancer regulates accessibility and RNAP binding, not only at specific localized sites but across the entire domain. The results support a facilitated tracking model for enhancer activity.
RNAP binding at ‘real’ genes and across the intergenic regions is qualitatively different. To demonstrate this, we used naturally occurring single-nucleotide polymorphisms (SNPs) to investigate allelic differences between binding of RNAP and activation of gene expression in wild-type cells and in cells carrying enhancer deletions or insulator insertion mutations. RNAP binding across the Igf2 and H19 genes is both enhancer-dependent and insulator-sensitive; that is, a functional insulator located between an enhancer and its regulated gene prevents RNAP binding and, likewise, prevents RNA transcription. Across the intergenic regions, RNAP binding and RNA transcription are similarly enhancer-dependent (see above). However, intergenic RNAP binding and transcription are not insulator-sensitive. The results indicate that insulators do not serve solely as a physical block for RNAP progression, but rather they specifically interfere with certain RNAP states or activities.
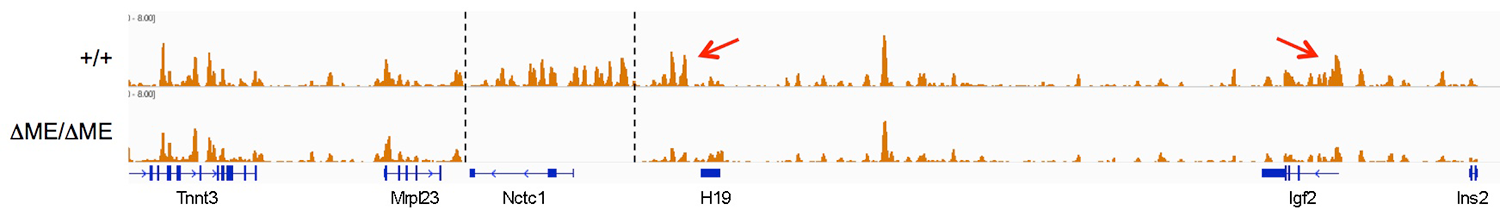
The muscle enhancer regulates RNAP binding and RNA transcription, but does not establish chromatin structures, because both RNA transcription and RNAP binding across the Igf2/H19 domain are entirely dependent upon the muscle enhancer. For example, levels of H19 RNA are reduced more than 10,000-fold in muscle cells in which the enhancer has been deleted. To test the dependence of chromatin structure on enhancer activity, we performed ChIP-seq on wild-type and on enhancer-deletion cell lines using antibodies against the histones H3K4me1, H3K43me3, and H3K36me3. Surprisingly, we saw no changes in the patterns of chromatin modification (Figure 4). Thus, a functional enhancer and active RNA transcription are not important for establishing chromatin structures at the Igf2/H19 domain.
To confirm and extend these observations, we are performing Cut&Run and ChIP assays to compare chromatin structures in wild-type cells and in cells lacking the ICR, the ME, and in cells in which we deleted CTCF sites associated with the muscle enhancer.
Functions of H19 lncRNA in regulating cell-cycle progression and senescence
To determine the biochemical functions of H19 lncRNA, we use in vitro models, including primary myoblasts, C2C12 myoblasts, and NIH3T3 cells. Abrupt depletion of H19 by either siRNA (small interfering RNA) or cre-induced recombination of H19–floxed alleles results in increased p21 RNA (p21 is a cyclin-dependent kinase inhibitor involved in cell-cycle arrest) and peptide, and such increased p21 activity in turn prevents cell-cycle progression and induces cellular senescence. H19 lncRNA regulation of p21 is at the level p21 mRNA stability and translation efficiency and occurs via the p21 3′ UTR. Genetic and biochemical analyses suggest that H19 lncRNA facilitates interactions between p21 mRNA and Wig-1 (which is involved in the regulation of mRNA stability). Current experiments focus on identifying the molecular mechanisms for these regulatory actions.
Functions of H19 lncRNA in regulating cardiac development
The Beckwith-Wiedemann syndrome (BWS) is a developmental disorder characterized by generalized overgrowth of the fetus and a high risk for several neonatal cancers. Many BWS patients also display cardiac problems. BWS can be explained by one of two different genetic lesions: loss of function of the CDKN1C gene or maternal loss of imprinting at the H19/Igf2 locus. Maternal loss of imprinting has the effect of doubling Igf2 expression while concomitantly reducing H19 RNA levels. Curiously, children born via artificial reproductive technology (ART) show increased incidence of BWS, which can be explained by increasingly frequent loss of H19/Igf2 imprinting in such children. Moreover, the children show high frequency of cardiac dysfunction. Taken together, such findings suggest that abnormal expression of the H19/Igf2 locus can lead to cardiac problems.
We observed that our BWS mouse model also results in cardiac dysfunction, as measured by echocardiography and ECG analyses. Molecular and molecular-genetic analyses demonstrate that biallelic Igf2 expression and loss of H19 play independent and distinct roles in generating the BWS phenotype. Biallelic expression of Igf2 results in elevated levels of circulating IGF2 peptide, which super-activates insulin and insulin-like receptor kinases in cardiomyocytes, resulting in hyper-activation of AKT/mTOR signaling pathways, which in turn causes cardiomyocyte hypertrophy and hyperplasia. Such effects result in a cardiac hypertrophy that is non-pathologic and transient, i.e., the hearts function normally and, as long as H19 levels are normal, the heart size normalizes after birth once Igf2 expression is repressed. Thus, there are no significant health effects associated with loss of imprinting of Igf2 only.
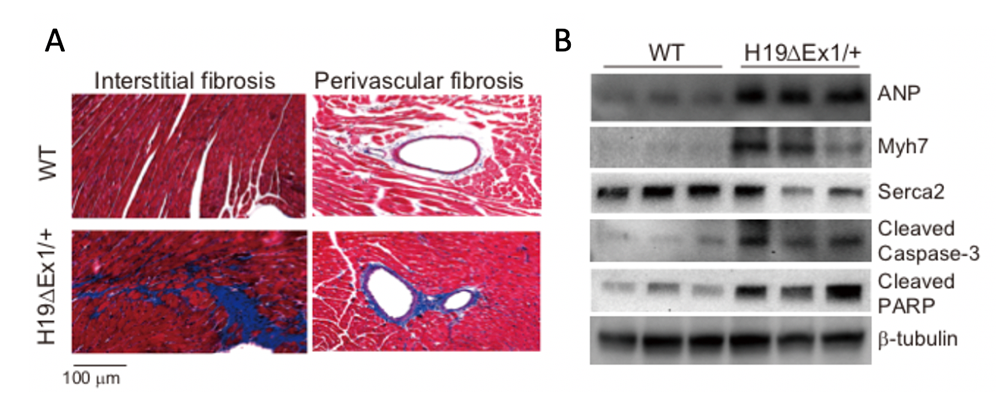
Loss of expression of H19 is pathologic (Figure 5). Hearts show progressive heart disease, manifested by hypertrophy, increased fibrosis, expression of cardiac failure markers, and reduced and abnormal heart function, as measured by echocardiography. H19 expression in hearts is restricted to endothelial cells. In vivo analyses of whole hearts and in vitro analyses of isolated endothelial cells show that reduction in H19 results in increased endothelial-to-mesenchymal transition (EMT). EMT is an essential feature of normal cardiac development; for example, formation of cardiac valves requires EMT. However, elevated frequency of EMT is associated with heart disease. Our data support the notion that H19 regulates the cell fate of endothelial cells, and future experiments aim to identify the underlying molecular mechanisms.
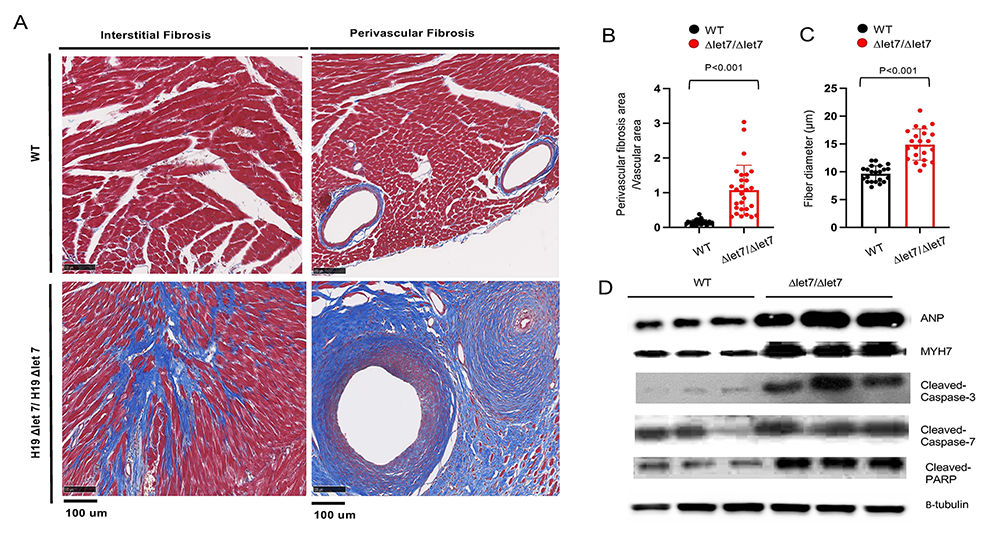
The H19 gene does not encode a protein. Rather H19 RNA is the functional gene product. The H19 precursor RNA is processed into two final products: a 2.3 kb lncRNA and microRNA 675. To determine which H19 RNA is essential for preventing cardiomyopathy, we developed two new mouse strains that selectively disrupt accumulation of the lncRNA or the miRNA. Disruption of the lncRNA is necessary and sufficient to induce cardiac disease. To understand the molecular bases for H19 lncRNA function, we generated alleles that disrupt specific domains. We observed that disruption of the let-7 microRNA (a microRNA highly conserved across species that controls stem-cell division and differentiation) binding sites on the H19 RNA results in the same myopathies as complete gene ablation (Figure 6). This was somewhat paradoxical given that increased let-7 function (which is expected when H19 is removed) is generally associated with protection from cardiomyopathy.
In hearts, H19 is not expressed in cardiomyocytes but only in a specific subset of endothelial cells. We combined a Tie2-Cre transgene and a conditional (floxed) H19 allele to delete H19 specifically in endothelial cells (and therefore deplete all H19 expression in the heart). We were surprised that this deletion did not cause cardiomyopathy, indicating that H19 expression outside the heart was responsible for preventing heart disease. Apart from endothelial cells, adult mice express H19 only in skeletal muscle. We therefore speculate that H19 expression in skeletal muscle is what prevents heart disease. We are currently investigating the role of skeletal muscle H19 in regulating levels of circulating musclin as an explanation for H19–dependent cardiomyopathy.
Function of H19 RNAs in muscle development
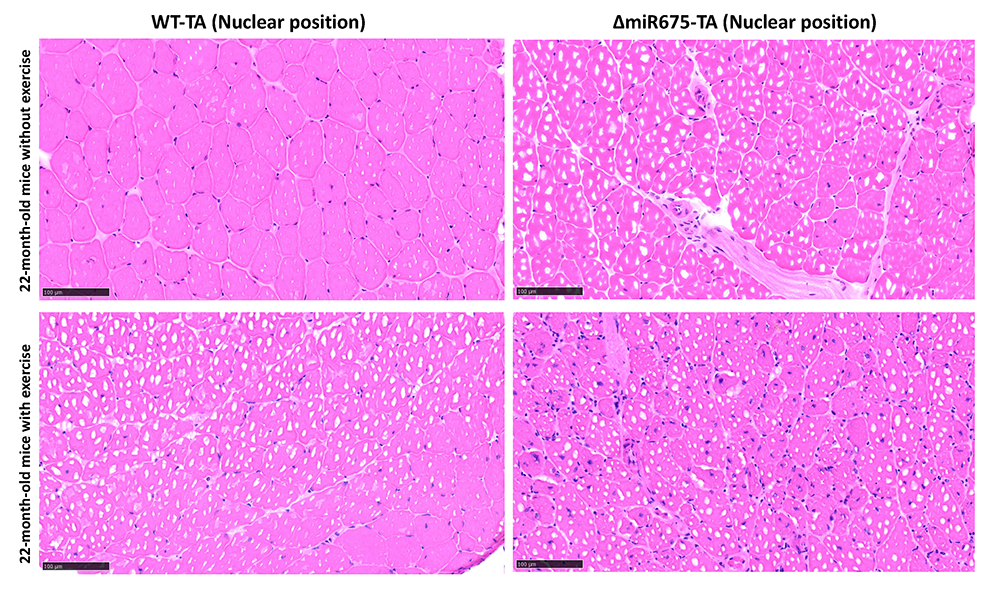
Studies in mice and humans indicate that H19 gene function is critical for normal development and function of skeletal muscle. As described in the previous section, H19 precursor RNA is processed into two distinct products: 2.3 kb lncRNA and a small microRNA (mi675). We generated mouse models that specifically ablate one or the other. The H19DeltaMIR allele generates an H19 lncRNA with a 40 bp deletion that is expressed at normal levels and maintains all H19 lncRNA functions that we tested. For example, the H19DeltaMIR lncRNA is sufficient to prevent cardiomyopathy and cellular senescence in myoblasts. However, H19DeltaMIRH19DeltaMIR mice present defects in muscle development and muscle regeneration. Specifically, muscle and muscle fibers are significantly smaller in mutant mice, and mutant mice cannot recover efficiently from muscle injury (Figure 7). The latter phenotype is exacerbated by age, indicating that H19 mutant mice are a model for sarcopenia. Current research focuses on identifying the molecular bases for these phenotypes.
Reducing Wapl dosage partially corrects embryonic growth and brain transcriptome phenotypes in Nipbl+/– embryos.
Cohesin rings interact with DNA to organize chromosomes into DNA–loop domains that modulate expression of thousands of genes. Cohesin is loaded onto chromosomes by NIPBL (a protein that is required for the association of cohesin with DNA) and unloaded by the cohesin-release factor WAPL, an essential regulator of chromatin structure and chromosome segregation. Thus, NIPBL and WAPL proteins have opposing biochemical activities with regard to cohesin loading and loop formation: less NIPBL means shorter and fewer loops and less WAPL means longer loops.
Haploinsufficiency for NIPBL leads to the Cornelia-de-Lange syndrome (CdLS), a developmental disorder. CdLS is highly pleiotropic but typically causes severe physical, cognitive, and medical challenges. CdLS is successfully modeled in Nipbl+/– mice. Nipbl+/– mice phenocopy the severe developmental and medical problems seen in CdLS patients and typically die perinatally.
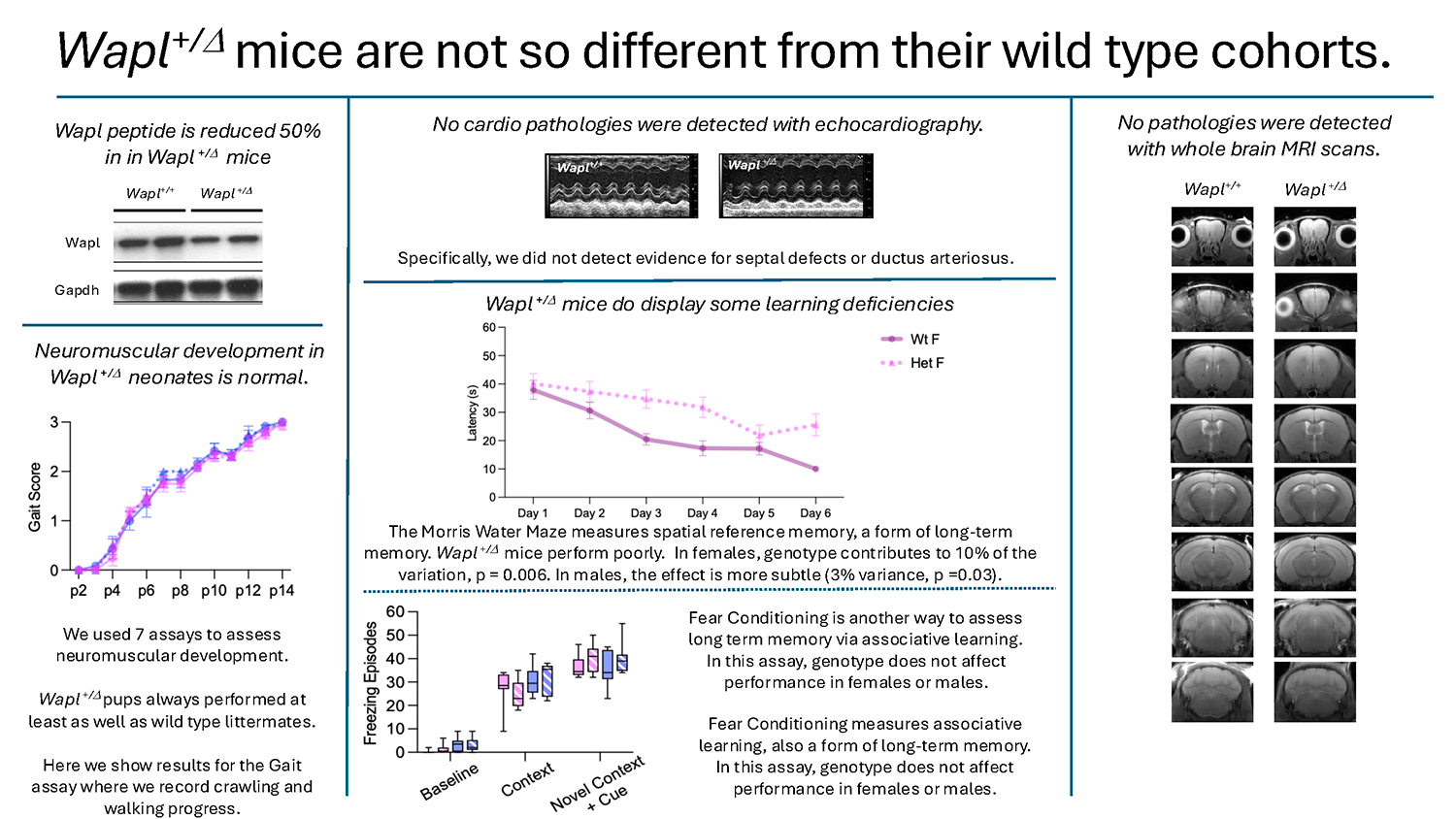
The impact of WAPL haploinsufficiency has been less well characterized. We generated Wapl+/– mice and are phenotyping these animals in consultation with collaborators at Boston Children’s Hospital, who are identifying and characterizing Wapl+/–patients. The impact of Wapl deficiency is limited. Wapl+/–mice are viable and fertile. Their development is normal. On several tests of muscle development and strength, they actually outperform wild-type littermates. In cognition assays, Wapl+/– mice display discrete phenotypes. Female Wapl+/– mice perform poorly on the Morris Water Maze (a test of long-term spatial learning) but perform at least as well as wild-type mice on tests for anxiety, short-term spatial memory, and long-term associative memory.
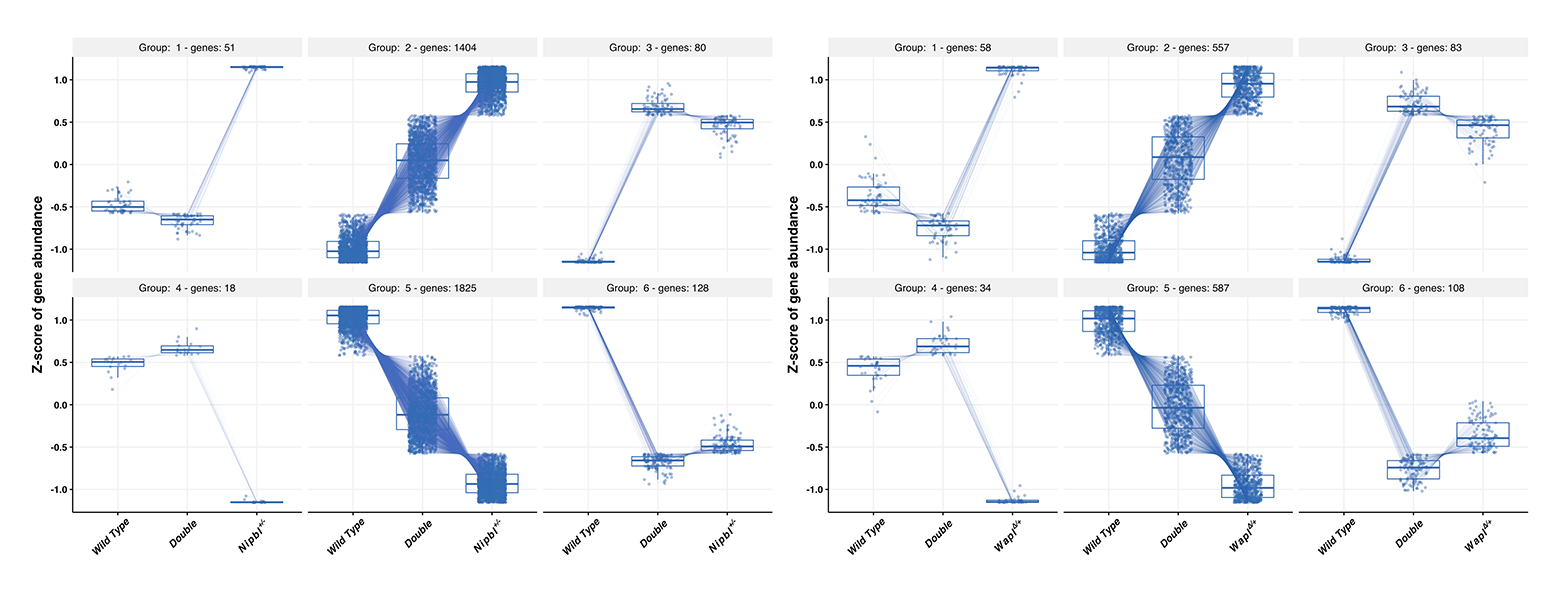
In Drosophila, reduced Nipbl function causes developmental defects. Importantly, in Drosophila, reducing Wapl and Nipbl function together alleviates many of the Nipbl–dependent developmental defects. To determine whether this is true in mammals, we generated the following mice: Wapl+/+ Nipbl+/+ (wild-type), Wapl+/– Nipbl+/+ (Wapl hets), Wapl+/+ Nipbl+/– (Nipbl hets), and Wapl+/– Nipbl+/– (double hets, i.e., two heterozygous gene pairs) littermates. Disruption of Wapl and Nipbl each lead to reduced growth and dysregulation of a (mostly) overlapping set of genes. Interesting, these phenotypes are not exacerbated but instead are partially rescued in the double mutant (Figure 8). The results suggest that Nipbl/Wapl balance (i.e., loading/unloading balance) is the critical factor in regulating cohesin function, and also suggest novel therapeutic approaches for CdLS patients.
Figure 9. Rescue of transcriptome phenotypes in Wapl+/– Nipbl+/– double heterozygotes
A. 94% of 3,506 genes dysregulated in Nipbl+/– embryonic brains are partially or fully rescued by reducing Wapl gene function.
B. Similarly, 87% of 1,427 genes dysregulated in Wapl+/– embryonic brains are rescued by reducing Nipbl function.
Figure 9. Rescue of transcriptome phenotypes in Wapl+/– Nipbl+/– double heterozygotes
A. 94% of 3,506 genes dysregulated in Nipbl+/– embryonic brains are partially or fully rescued by reducing Wapl gene function.
B. Similarly, 87% of 1,427 genes dysregulated in Wapl+/– embryonic brains are rescued by reducing Nipbl function.
Publications
- Decreasing Wapl dosage partially corrects embryonic growth and brain transcriptome phenotypes in Nipbl+/- embryos. Sci Adv 2022 8:eadd4136
- Cardiac pathologies in mouse loss of imprinting models are due to misexpression of H19 long noncoding RNA. eLife 2021 10:e67250
- Conditional ablation and conditional rescue models for Casq2 elucidate the role of development and of cell-type specific expression of Casq2 in the CPVT2 phenotype. Hum Mol Genet 2018 27:1533–1544
- Loss of imprinting mutations define both distinct and overlapping roles for misexpression of IGF2 and of H19 lncRNA. Nucleic Acids Res 2017 45:12766–12779
- Circular RNAs and competing endogenous (ceRNA) networks. Transl Cancer Res 2018 7:S624–628
Collaborators
- Philip Boone, MD, PhD, Boston Children's Hospital, Boston, MA
- Danielle A. Springer, VMD, DACLAM, Murine Phenotyping Core Facility, NHLBI, Bethesda, MD
Contact
For more information, email kpfeifer@helix.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/pfeifer.