Regulatory Small RNAs and Small Proteins
- Gisela Storz,
PhD, Head, Section on Environmental Gene Regulation - Aixia Zhang, PhD, Staff Scientist
- Maciej M. Basczok, PhD, Postdoctoral Fellow
- Aisha Burton Okala, PhD, Postdoctoral Fellow
- Rajat Dhyani, PhD, Postdoctoral Fellow
- Chelsey R. Fontenot, PhD, Postdoctoral Fellow
- Shuwen Shan, PhD, Postdoctoral Fellow
- Narumon Thongdee, PhD, Postdoctoral Fellow
- Rilee D. Zeinert, PhD, Postdoctoral Fellow
- Aoshu Zhong, PhD, Postdoctoral Fellow
- Dennis X. Zhu, PhD, Postdoctoral Fellow
- Miranda Alaniz, BS, Postbaccalaureate Fellow
- Amanda P. Brewer, BS, Postbaccalaureate Fellow
- Anna J. Bryant, BS, Postbaccalaureate Fellow
- Juwaan Douglas-Jenkins, BS, Postbaccalaureate Fellow

The group currently has two main interests: identification and characterization of small noncoding RNAs (sRNAs), and identification and characterization of small proteins of less than 50 amino acids. Both small RNAs and small proteins have been overlooked because they are not detected in biochemical assays, and the corresponding genes are missed by genome annotation and are poor targets for genetic approaches. However, both classes of small molecules are being found to have important regulatory roles in organisms ranging from bacteria to humans.
Identification and characterization of small regulatory RNAs
During the past 20 years, we have carried out several different systematic screens for small regulatory RNAs in Escherichia coli. The screens included computational searches for conservation of intergenic regions and direct detection after size selection or co-immunoprecipitation with RNA–binding proteins. Most recently, we have been using deep sequencing approaches to map the 5′ and 3′ ends of all transcripts to further extend our identification of small RNAs in a range of bacteria species [Reference 1], work that showed that sRNAs are encoded by diverse loci, including sequences overlapping mRNAs.
A major focus for the group has been to elucidate the functions of the small RNAs that we and others identified. Early on, we showed that the OxyS RNA, whose expression is induced in response to oxidative stress, acts to repress translation through limited base pairing with target mRNAs. We discovered OxyS action is dependent on the Sm–like Hfq protein, which acts as a chaperone to facilitate OxyS RNA base pairing with its target mRNAs. Follow-up studies allowed us to learn more about the mechanism by which the Hfq protein facilitates base pairing through multiple RNA binding domains. In a recent collaboration with the group of Susan Gottesman, we found that an acetyltransferase has a moonlighting function: binding to and modulating the RNA–binding repertoire of Hfq [Reference 2]. We also started to explore the role of ProQ, a second RNA chaperone in E. coli and, by comparing the sRNA–mRNA interactomes by deep sequencing, found that ProQ and Hfq have overlapping as well as competing roles in the cell. It is likely that still other RNA–binding proteins, such as KH–domain (nucleic acid recognition motifs) proteins, are involved in small RNA–mediated regulation.
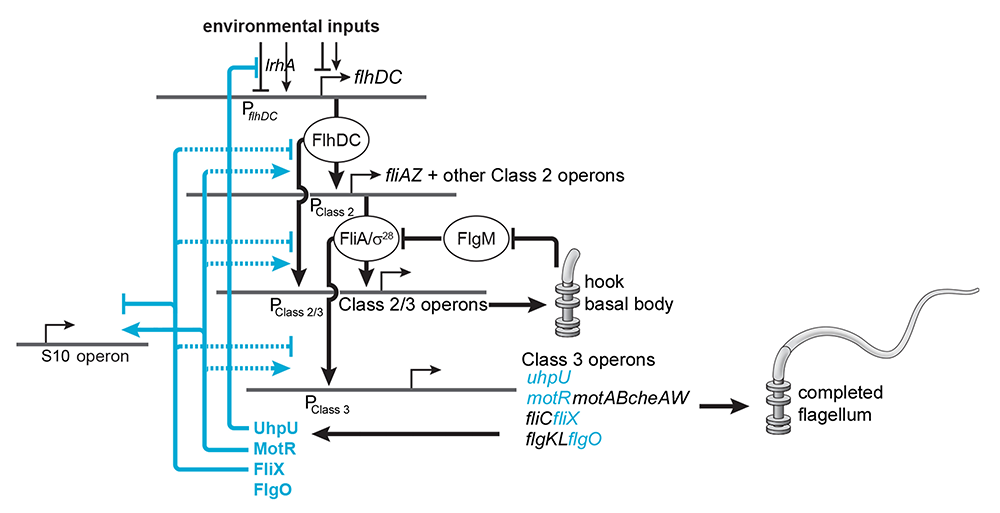
Hfq–binding small RNAs, which act through limited base pairing, are integral to many different stress responses in E. coli and other bacteria, as well as during the interaction between bacteria and bacteriophages. Studies on the Hfq–binding sRNAs has given insights into the nuanced control of the regulatory networks as well as into bacterial physiology in general [Reference 3]. For example, we showed that the Spot 42 RNA, whose levels are highest when glucose is present, plays a broad role in catabolite repression by directly repressing genes required for the consumption of diverse non-preferred carbon sources. Similarly, we found that a small RNA derived from the 3′ UTR of the glnA encoding glutamine synthetase impacts E. coli growth under low nitrogen conditions by modulating the expression of genes that affect carbon and nitrogen flux. In recent work, we described four UTR–derived sRNAs (UhpU, MotR, FliX, and FlgO), whose expression is controlled by the flagella sigma factor σ28 (fliA) and which have varied effects on flagellin protein levels, flagella number, and cell motility [Reference 4]. Intriguingly, MotR, corresponding to the 5′ UTR of an early gene in the flagella regulon, activates flagellar synthesis, while FliX, corresponding to a late gene in the flagella regulon, downregulates flagellar synthesis, illustrating how sRNA–mediated regulation can overlay a complex network enabling temporal control (Figure 1). As more and more sRNAs encoded by 5′ or 3′ UTRs or internal to coding sequences are being found, our observations raise the possibility that phenotypes currently attributed to protein defects result from deficiencies in unappreciated regulatory RNAs.
Figure 1. Network of sRNAs controlling flagella synthesis
σ28-dependent sRNAs control flagella synthesis at different levels. UhpU activates the flagellar regulon by repressing a regulator of flhDC. MotR and FliX, respectively, activate (arrow) and repress (bar) middle and the late gene expression. Thus, MotR acts as an accelerator and FliX as a decelerator of flagella synthesis. Solid lines indicate documented base pairing between the sRNA and mRNA, and dotted lines indicate that the exact mechanism of regulation is not known.
Figure 1. Network of sRNAs controlling flagella synthesis
σ28-dependent sRNAs control flagella synthesis at different levels. UhpU activates the flagellar regulon by repressing a regulator of flhDC. MotR and FliX, respectively, activate (arrow) and repress (bar) middle and the late gene expression. Thus, MotR acts as an accelerator and FliX as a decelerator of flagella synthesis. Solid lines indicate documented base pairing between the sRNA and mRNA, and dotted lines indicate that the exact mechanism of regulation is not known.
One interesting recent observation is that some small RNAs have dual functions in that they act by both base pairing and by encoding a small, regulatory protein [Reference 5]. For example, we discovered the Spot 42 RNA also encodes a 15–amino acid protein (denoted SpfP). Overexpression of just the small protein or just the base pairing activity both prevented growth on galactose, revealing that the small protein and the small RNA impact the same pathway. As a second example, we found a 164–nucleotide RNA previously shown to encode a 28–amino acid protein (denoted AzuC) also base pairs with the cadA and galE mRNAs to block expression. Interestingly, AzuC translation interferes with the observed repression of cadA and galE by the RNA, and base pairing interferes with AzuC translation, demonstrating that the translation and base-pairing functions compete. We hypothesize that many more dual-function RNAs remain to be discovered and suggest that they can be exploited to control gene expression at many levels. We successfully constructed a functional synthetic dual-function regulator from a small protein and a small protein encoded by adjacent genes and used this synthetic construct to study the functional organization of dual-function RNAs.
In addition to small RNAs that act via limited base pairing, we have been interested in regulatory RNAs that act by other mechanisms. For instance, early work showed that the 6S RNA binds to and modulates RNA polymerase by mimicking the structure of an open promoter. In another study, we discovered that the broadly conserved yybP–ykoY riboswitch motif, which is found in the 5′ UTR of the mntP gene encoding a manganese exporter, directly binds manganese, resulting in a conformation that liberates the ribosome-binding site, thereby increasing MntP synthesis.
Further studies to characterize other Hfq– and ProQ–binding RNAs and their physiological roles and evolution, as well as regulatory RNAs that act in ways other than base pairing, are ongoing.
Identification and characterization of small proteins
In our genome-wide screens for small RNAs, we found that a number of short RNAs actually encode small proteins. The correct annotation of the smallest proteins is one of the biggest challenges of genome annotation. Further, there is limited evidence that proteins are synthesized from annotated and predicted short ORFs. Although these proteins have largely been missed, the few small proteins that have been studied in detail in bacterial and mammalian cells have been shown to have important functions in regulation, signaling, and in cellular defenses [Reference 6]. We thus established a project to identify and characterize proteins of less than 50 amino acids.
We first used sequence conservation and ribosome binding–site models to predict genes encoding small proteins of 16–50 amino acids in the intergenic regions of the model E. coli genome. We tested expression of these predicted, as well as previously annotated, small proteins by integrating the sequential peptide affinity tag directly upstream of the stop codon on the chromosome and assaying for synthesis using immunoblot assays. The approach confirmed the synthesis of 20 previously annotated and 18 newly discovered proteins of 16–50 amino acids. We also carried out a complementary approach based on genome-wide ribosome profiling of ribosomes arrested on start codons to identify many additional candidates; the synthesis of 38 of these small proteins again was confirmed by chromosomal tagging. These studies, together with the work of others, have documented that E. coli synthesize over 150 small proteins. Systematic assays for the accumulation of tagged versions of the proteins showed that many small proteins accumulate under specific growth conditions or after exposure to stress. Many small proteins also are predicted to consist of a single transmembrane alpha-helix and have been found to be in the inner membrane. However, despite their diminutive size, small membrane proteins display considerable diversity in topology and insertion pathways.
To elucidate the functions of the small proteins, we are using the tagged derivatives and information about synthesis and subcellular localization, along with many of the approaches the group has used to characterize the functions of small regulatory RNAs. The combined approaches are providing insights into small-protein actions, which are relevant to all organisms. For example, we discovered that the 49–amino acid inner-membrane protein AcrZ, whose synthesis is increased in response to noxious compounds such as antibiotics and oxidizing agents, associates with the inner-membrane AcrB component of the AcrAB-TolC multidrug, resistance-nodulation-division (RND) efflux pump. Mutants lacking AcrZ are sensitive to many, but not all, of the antibiotics transported by AcrAB-TolC as a consequence of AcrZ–imposed changes on the conformation of AcrB drug-binding pockets. We also found that synthesis of a 42–amino acid protein MntS is repressed by high levels of manganese by the MntR transcription factor. The lack of MntS leads to decreased activities of manganese-dependent enzymes under manganese-poor conditions, while overproduction of MntS leads to very high intracellular manganese and bacteriostasis under manganese-rich conditions. These and other phenotypes led us to propose that MntS modulates intracellular manganese levels by inhibiting the manganese exporter MntP. Additionally, we showed that the 31–amino acid inner-membrane protein MgtS, whose synthesis is induced by very low magnesium by the PhoPQ two-component system (a transcriptional regulator), acts to increase intracellular magnesium levels and maintain cell integrity upon magnesium depletion. Upon development of a functional, tagged derivative of MgtS, we found that MgtS interacts with MgtA to increase the levels of this P-type ATPase magnesium transporter under magnesium-limiting conditions. MgtS stabilization of MgtA provides an additional layer of regulation of this tightly controlled magnesium transporter. RND efflux pumps and P-type ATPase transporters are broadly distributed, and we suggest that many more members will be found to be regulated by small proteins.
The ribosome profiling used to identify the intergenic-encoded small proteins revealed that there is significant translation initiation within larger open reading frames in the E. coli genome. All five E. coli genes encoding Rpn (recombination-promoting nuclease) proteins have such an internal translation site. We showed that the small, highly variable Rpn C-terminal domains (RpnS), which are translated separately from the full-length proteins (RpnL), directly block the activities of the toxic full-length RpnL proteins, comprising a novel toxin-antitoxin system [Aoyama JJ, Storz G. Trends Biochem Sci 2023;48:1035–1043]. The crystal structure of RpnAS revealed a dimerization interface encompassing a helix that can have four amino acid repeats, whose number varies widely among strains of the same species. Consistent with strong selection for the repeat variation, we documented that plasmid-encoded RpnP2L protects E. coli against specific phages. We propose that intragenic-encoded small proteins that serve regulatory roles remain to be discovered in all organisms.
The ribosome profiling also revealed that some regulatory RNAs also encode a small protein and are thus dual-function RNAs [Reference 5]. As mentioned above, we documented that the 109–nucleotide Spot 42 RNA, which is one of the best characterized base-pairing small RNAs (sRNAs) in E. coli, encodes a 15–amino acid protein (denoted SpfP). Co-purification experiments revealed that SpfP binds to the global transcriptional regulator CRP. The binding blocks the ability of CRP to activate specific genes, impacting the kinetics of induction when cells are shifted from glucose to galactose medium. Thus, the small protein reinforces the feedforward loop regulated by the base-pairing activity of the Spot 42 RNA. Another 164–nucleotide RNA was previously shown to encode a 28–amino acid, amphipathic-helix protein (denoted AzuC) but later shown to also act by base pairing (AzuR). We discovered that the membrane-associated AzuC protein interacts with GlpD, the aerobic glycerol-3-phosphate dehydrogenase, and increases dehydrogenase activity by recruiting the protein to the membrane.
Our work, along with related findings by others in eukaryotic cells, supports our hypothesis that small proteins are an overlooked but important class of proteins, which we continue to study.
Additional Funding
- NICHD Early Career Award
- NIGMS Postdoctoral Research Associate (PRAT) Program
- Scientific Director's Award 2023–2024
Publications
- Extensive diversity in RNA termination and regulation revealed by transcriptome mapping for the Lyme pathogen Borrelia burgdorferi. Nat Commun 2023 14:3931
- An acetyltransferase moonlights as a regulator of the RNA binding repertoire of the RNA chaperone Hfq in Escherichia coli. Proc Natl Acad Sci USA 2023 120:e2311509120
- Insights into bacterial metabolism from small RNAs. Cell Chem Biol 2024 31:1571–1577
- σ28-dependent small RNA regulation of flagella biosynthesis. eLife 2023 12:RP87151
- Large roles of small proteins. Annu Rev Microbiol 2024 78:1–22
- Toxic antiphage defense proteins inhibited by intragenic antitoxin proteins. Proc Natl Acad Sci USA 2023 120:e2307382120
Collaborators
- Ryan K. Dale, PhD, Bioinformatics and Scientific Programming Core, NICHD, Bethesda, MD
- Fred Dyda, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
- Caroline Esnault, PhD, Bioinformatics and Scientific Programming Core, NICHD, Bethesda, MD
- Susan Gottesman, PhD, Laboratory of Molecular Biology, Center for Cancer Research, NCI, Bethesda, MD
- Alison B. Hickman, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
- Aravind Iyer, PhD, NLM/NCBI, NIH, Bethesda, MD
- Michael Jarnik, PhD, Division of Neurosciences and Cellular and Structural Biology, NICHD, Bethesda, MD
- Xiaofang Jiang, PhD, National Library of Medicine, NIH, Bethesda, MD
- Michael T. Laub, PhD, Massachusetts Institute of Technology, Cambridge, MA
- Nadim Majdalani, PhD, Laboratory of Molecular Biology, Center for Cancer Research, NCI, Bethesda, MD
- Mikolaj Olejniczak, PhD, Institute of Molecular Biology and Biotechnology, Adam Mickiewicz University, Poznan, Poland
- Kai Papenfort, PhD, Institute of Microbiology, Friedrich-Schiller-Universität, Jena, Germany
- Svetlana A. Shabalina, PhD, National Library of Medicine, NIH, Bethesda, MD
- Chin-Hsien Tai, PhD, Laboratory of Molecular Biology, Center for Cancer Research, NCI, Bethesda, MD
- Joseph T. Wade, PhD, Wadsworth Center, New York State Department of Health, Albany, NY
- Henry Zhang, PhD, Bioinformatics and Scientific Programming Core, NICHD, Bethesda, MD
Contact
For more information, email storzg@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/storz.