You are here: Home > Section on Neurophysiology and Biophysics
Glutamate Receptor Structural Biology

- Mark L. Mayer, PhD, Head, Section on Neurophysiology and Biophysics
- Charu Chaudhry, PhD, Postdoctoral Fellow
- Utpal Das, PhD, Visiting Fellow
- Carla Glasser, PhD, Technical Specialist
- Janesh Kumar, PhD, Visiting Fellow
- Andrew Plested, PhD, Visiting Fellow
- Yongneng Yao, PhD, Visiting Fellow
Ionotropic glutamate receptors (iGluRs) are membrane proteins that act as molecular pores and mediate signal transmission at the majority of excitatory synapses in the mammalian nervous system. The seven iGluR gene families in humans encode 18 subunits, which assemble to form three major functional families named after the ligands that were first used to identify iGluR subtypes in the late 1970s: AMPA, kainate, and NMDA. Given the essential role of iGluRs in normal brain function and development, and mounting evidence that dysfunction of iGluR activity mediates several neurological and psychiatric diseases and damage during stroke, we devote substantial effort to analyzing iGluR function at the molecular level. Atomic resolution structures solved by protein crystallization and X-ray diffraction provide a framework for designing electrophysiological and biochemical experiments aimed at defining the mechanisms underlying ligand recognition, the gating of ion channel activity, and the action of allosteric modulators. Information derived from these experiments will permit the development of subtype-selective antagonists and allosteric modulators with novel therapeutic applications and reveal the inner workings of a complicated protein machine that plays a key role in brain function.
Crystallographic and functional analysis of an allosteric binding site for sodium
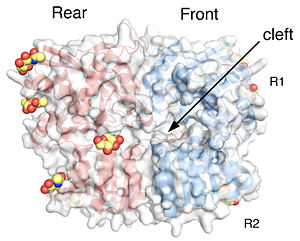
Click image to enlarge.
Crystal structure of the GluR6 ATD dimer assembly
The two subunits in a dimer assembly are shown as red and blue ribbon diagrams with a transparent molecular surface representation. Each subunit has an upper (R1) and lower (R2) domain, both of which are tightly apposed. Glycans that decorate the solvent-exposed surface and were trimmed to NAG molecules prior to crystallization are shown in CPK format.
Kainate-subtype glutamate receptors are strongly modulated by monovalent anions and cations. In the absence of either chloride or sodium, the receptors become nonfunctional. We used a combined experimental approach based on crystallography, patch-clamp recording, and all-atom molecular dynamics (MD) simulations to identify the binding site for sodium and the mechanism by which sodium modulates kainate receptor activity. We solved structures for the GluR5 ligand–binding domain dimer complex with lithium, sodium, potassium, rubidium, cesium, and ammonium ions in the cation-binding site. We observed two sodium binding sites in a dimer assembly (one per subunit); they flank the previously identified anion-binding site that lies on the molecular two-fold axis of symmetry. Sodium stabilizes the dimer assembly in its active conformation, which is required for ion channel gating; in the absence of sodium, the receptors desensitize much faster. Sodium selectivity is conferred by a high electric field strength in the cation-binding site, but larger cations can bind with lower affinity. Functional studies show that the cation-binding site is allosterically coupled to the anion-binding site. All-atom MD simulations and free energy calculations reveal that the binding of chloride is favored by 3 to 5 kcal per mole when the cation-binding site is occupied by sodium. Mutational analysis and molecular modeling demonstrated that it is possible to convert the sodium-binding site to a site with micromolar affinity for the divalent cations calcium and magnesium, namely by substituting an aspartate residue for a hydrophobic amino acid that caps the sodium-binding site in kainate receptors. AMPA receptors, which are insensitive to allosteric modulation by either sodium or calcium, harbor a lysine at this site. Amino acid sequence analysis indicated that the divergence between iGluRs with and without allosteric binding sites for sodium arose early in evolution. We were unable to crystallize a kainate receptor with the aspartate mutation and therefore made molecular models of the binding site. The model reveals a unique geometry, with three closely apposed carboxylate groups together with two backbone carbonyl oxygen atoms that provide ligands for binding to calcium similar to those found in the protein data bank for a diverse range of proteins with calcium binding sites.
Molecular biophysical studies on kainate receptor dimer assembly
For kainate-, but not AMPA-subtype glutamate receptors, the binding of sodium and chloride ions to discrete, electrostatically coupled sites in the extracellular ligand-binding domain (LBD) dimer assembly regulates the rate of entry into the desensitized state, which occurs when the dimer interface ruptures and the channel closes. Studies on glutamate receptors have defined the LBD dimer assembly as a key functional unit that controls activation and desensitization.
Direct measurement of the effects of allosteric ions on dimer assembly by kainate-subtype iGluRs is not possible with electrophysiological techniques. To obtain proof that allosteric ions regulate dimer formation, we developed a series of GluR6 dimer interface mutations remote from the ion-binding sites and set out to develop a preparation amenable to analysis by analytical ultracentrifugation (AUC). We designed the mutants on the basis of crystal structures for wild-type GluR5 and GluR6 dimers, using electrophysiological analysis to search for a phenotype with slowed kinetics of desensitization. We prepared the isolated ligand-binding domains of the same mutants and measured their affinity for dimer formation by sedimentation velocity analysis over a range of protein concentrations and by sedimentation equilibrium. The most stable mutants increased dimer stability by at least 3 kcal per mole, with excellent agreement between the Kd for dimer assembly and the rate of desensitization measured in functional experiments. To explore the molecular basis for control of dimer assembly, we crystallized the wild-type GluR6 dimer together with a series of six mutants at resolutions of 1.2 to 1.5 Å. Analysis of these structures and of the effects of allosteric ions on dimer assembly measured by AUC reveals that the dimer interface of kainate receptors is intrinsically less stable than that for AMPA receptors.
We then used AUC to directly measure the energetic effects of allosteric ions on kainate receptor dimer stability in solution, using a GluR6 mutant that desensitizes slowly. Our results showed that sodium and chloride ions modulate kainate receptor dimer affinity as much as 50-fold and that removal of either chloride or sodium disrupts the dimer. The applicability of a similar allosteric mechanism for modulation of delta2 glutamate receptors by calcium was also tested. Our results indicate that ions can contribute substantial free energy to active-state stabilization in both these receptors and provide quantitative measurements of the energetic consequences of allosteric ion binding to a ligand-gated ion channel. We propose that this difference arose during evolution to accommodate the allosteric modulatory effects of sodium and chloride ions on dimer stability, which are unique to the kainate receptor gene family.
Structural studies on the amino-terminal domain of iGluRs
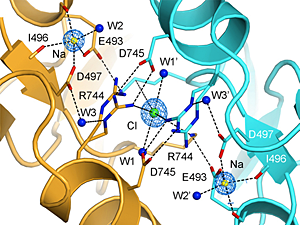
Click image to enlarge.
Crystal structure of the allosteric ion binding sites in a GluR6 ligand binding domain dimer
The two subunits in a dimer assembly are shown using ribbon diagrams colored gold and cyan, respectively. Fo-Fc omit electron density maps show the location of a single chloride ion on the dimer 2-fold axis of molecular symmetry, flanked by a pair of sodium ions. Side chains that coordinate the ions are drawn in stick representation and, for Arg744, show alternate conformations, reflecting instability of the Arg744-Asp745 salt bridge, as found also in MD simulations.
Glutamate receptor ion channels are multidomain membrane proteins that assemble from tetramers of approximately 440 kD. Numerous crystal structures have been solved for the ligand-binding domains, which have a molecular weight of approximately 30 kD per subunit—approximately one-quarter of the mass of an intact receptor. Extensive trials with bacterial expression systems, which with one exception have been used for all published ligand-binding domain structures, failed to produce monodisperse soluble protein for other iGluR domains. The 400 amino acid amino-terminal domain (ATD) is an important structural target because it controls subtype-selective assembly in native iGluRs, limiting assembly to members of the same functional family. Protein expression in mammalian cells at levels sufficient for structural biology is much more difficult than expression in E. coli, but has the advantages that several check points select for correctly folded proteins and add sugars and other post-translational modifications required for normal function. Although a variety of cell-biological and biochemical techniques are required to subsequently trim the sugar chains, in order to obtain proteins that crystallize and diffract to high resolution, and the yields are lower than for prokaryotic expression, mammalian cell culture is the only current approach likely to succeed for studies of the ATD. In ongoing work, we have screened the ATDs from several iGluR subtypes for expression in mammalian cells, and for GluR6 ATD we obtain around 4 mg/l of purified protein. We have performed sedimentation experiments using an analytical ultracentrifuge, and crystallization trials using a nanoliter pipetting robot. Diffraction data to a resolution of 2.7 Å for a complete data set was collected at APS and the structure refined to good statistics.
The GluR6 ATD forms dimers in solution at micromolar protein concentrations and crystallizes as a dimer. Unexpectedly, each subunit adopts an intermediate extent of domain closure compared with the apo and ligand-bound complexes of LIVBP (leucine-isoleucine-valine binding protein) and G protein–coupled glutamate receptors (mGluRs), and the dimer assembly has a markedly different conformation from that found in mGluRs. This conformation is stabilized by contacts between large hydrophobic patches in the R2 domain that are absent from NMDA receptors, suggesting that the ATDs of individual glutamate receptor ion channels have evolved into functionally distinct families. Crystallographic symmetry operations reveal the assembly mechanism within a dimmer-of-dimers assembly and indicate that the pairs of dimers assemble via limited contacts on the lateral edges of domain R2 to generate a W-shaped structure.
Structural analysis of NR3 ligand binding selectivity
NR3-subtype glutamate receptors have a unique developmental expression profile but are the least characterized members of the NMDA receptor gene family; those members play key roles in synaptic plasticity and brain development. Using ligand-binding assays, crystallographic analysis, and all-atom MD simulations, we investigated mechanisms underlying the binding of NR3A and NR3B to glycine and D-serine, which are candidate neurotransmitters for NMDA receptors containing NR3 subunits. The ligand-binding domains of both NR3 subunits adopt a similar extent of domain closure to that found in the corresponding NR1 complexes but have a unique loop 1 structure distinct from those in all other glutamate receptor ion channels. Within their ligand-binding pockets, NR3A and NR3B have strikingly different hydrogen bonding networks and solvent structures from those in NR1 and fail to undergo a conformational rearrangement observed in NR1 upon binding to the partial agonist ACPC. Replica exchange MD simulations of 650 ns duration revealed numerous interdomain contacts that stabilize the agonist-bound closed-cleft conformation as well as a novel twisting motion for the loop 1 helix that is unique in NR3 subunits. Mutation of these sites destabilized ligand binding as measured by titration assays that use quenching of endogenous tryptophan fluorescence.
Publications
- Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J 2008 27:2158-2170.
- Plested AJ, Vijayan R, Biggin P, Mayer ML. Molecular basis of kainate receptor modulation by sodium. Neuron 2008 58:720-735.
- Chaudhry C, Plested AJ, Schuck P, Mayer, ML. Energetics of glutamate receptor ligand binding domain dimer assembly is modulated by allosteric ions. Proc Natl Acad Sci USA 2009 106:12329-12334.
- Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6 subtype glutamate receptor ion channels. Nat Struct Mol Biol 2009 16:631-638.
- Chaudhry C, Weston MC, Schuck P, Rosenmund C, Mayer, ML. Stability of ligand binding domain dimer assembly controls kainate receptor desensitization. EMBO J 2009 28:1518-1530.
Collaborators
- Philip Biggin, PhD, University of Oxford, Oxford, UK
- David Jane, PhD, University of Bristol, Bristol, UK
- Christian Rosenmund, PhD, Baylor College of Medicine, Houston, TX
- Peter Schuck, PhD, Protein Biophysics Resource, DBEPS, NIH, Bethesda, MD



