You are here: Home > Section on Eukaryotic Transposable Elements
Retrotransposons as Models for the Replication of Retroviruses

- Henry L. Levin, PhD, Head, Section on Eukaryotic Transposable Elements
- Angela Atwood-Moore, BA, Senior Research Assistant
- Atreyi Chatterjee, PhD, Visiting Fellow
- Yujin Cui, BA, Volunteer
- Robin Cutler, BA, Postbaccalaureate Fellow
- Gang Feng, BA, Graduate Student
- Yabin Guo, PhD, Visiting Fellow
- Elizabeth Humes, BA, Postbaccalaureate Fellow
- Anusua Majumdar, PhD, Visiting Fellow
- Jung Min Park, BA, Postbaccalaureate Fellow
- Tracy Ripmaster, PhD, Research Assistant
Diseases such as AIDS and leukemia caused by retroviruses have intensified the need to understand the mechanisms of retrovirus replication. One of our objectives is to understand how retroviral cDNAs are integrated into the genome of infected cells. Because of their similarities to retroviruses, long terminal repeat (LTR)-retrotransposons are important models for retrovirus replication. The retrotransposon under study in our laboratory is the Tf1 element of the fission yeast Schizosaccharomyces pombe. We are particularly interested in Tf1 because its integration exhibits a strong preference for pol II promoters. This choice of target sites is similar to the strong integration preferences of human immunodeficiency virus 1 (HIV-1) and murine leukemia virus (MLV) for pol II transcription units. Currently, it is not clear how these viruses recognize their target sites and perform integration. We therefore study the integration of Tf1 as a model system with which we hope to uncover mechanisms general to the selection of integration sites. An understanding of the mechanisms responsible for targeted integration could lead to new approaches for antiviral therapies.
Determinants of the integration pattern of retrotransposon Tf1 in the fbp1 promoter of Schizosaccharomyces pombe
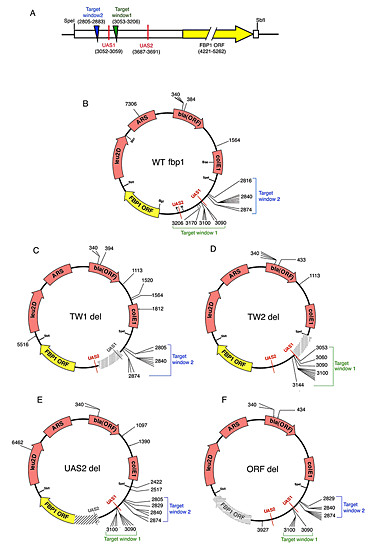
Figure 1. A plasmid-based assay for integration of Tf1 in fbp1
A. Linear diagram representing the fbp1 promoter in construct pHL2679. The construct has a 2.9kb fbp1 sequence that includes UAS1 (in red), UAS2 (in red) and the fbp1 ORF (in yellow block arrow). Integration occurred in TW1 (green triangle) and TW2 (blue triangle). In parentheses are the coordinate numbers based on the WT fbp1plasmid (pHL2679). The target plasmid assay was used to map integration in plasmids that contained WT fbp1(B), TW1 deleted (C), TW2 deleted (D), UAS2 deleted (E), and ORF deleted (F). The positions of integration events are shown as black lines with plasmid coordinate numbers based on the sequence of WT fbp1. The positions of TW1 and TW2 are shown for each plasmid. Restriction sites for BseYI (Bse), SpeI (SpeI), NgOMIV (N), PacI (P), BglII (Bgl), SbfI (SbfI) and BsrGI (Bsr) are shown. (click image to enlarge)
A specific goal of our research is to identify the mechanism that directs integration to regions containing pol II promoters. To study insertion patterns in specific genes, a target plasmid assay was developed. Integration into the promoter of fbp1clustered within 10 bp of a transcription enhancer called upstream activating sequence 1 (UAS1). Integration into the promoter of fbp1 depended on UAS1 sequence and Atf1p, a transcription activator that binds to UAS1. To identify the key determinants responsible for targeting integration in the fbp1 promoter, we conducted an extensive study of the promoter sequences (Majumdar et al, 2010). We found that two discrete target windows close to UAS1 were the only sequences in the promoter required for the pattern of integration (Figure 1). These two target windows functioned independently of each other, and each one was found to be sufficient to function as an efficient target of integration. Although Atf1p is necessary for directing integration to UAS1, it may be that, by activating transcription, Atf1p induces subsequent steps of transcription that are more directly responsible for directing integration. If the role of Atf1p in integration were indirect, other factors that promote fbp1 transcription would also influence integration at this promoter. However, other known factors that mediate fbp1 transcription—Pcr1p, Rst2p, and Tup11p/Tup12p—were found not to contribute to integration. UAS2 is an independent enhancer in the promoter of fbp1and was not a target of integration. Nevertheless, we found that UAS2 did promote efficient transcription of fbp1. In addition, we found a synthetic promoter induced by lexA fused to an activator, VP16, was not a target of Tf1 integration. The data indicate that transcription activity of a promoter is not sufficient to mediate integration but that Atf1p plays a direct and specific role in targeting integration to UAS1 of the fbp1 promoter.
The integrase of Tf1 interacts directly with the b-ZIP domain of Pcr1p.
The role of Atf1p in integration may be to bind to and recruit integrase to UAS1. To test integrase for direct interactions with Atf1p, we conducted pull-down experiments. Various domains of integrase and Atf1p were fused to epitope tags, and the recombinant proteins were purified from bacteria. The experiments demonstrated that the catalytic core of integrase interacted with the b-ZIP domain of Atf1p. While the in vitro results with recombinant proteins indicated that integrase and Atf1p are capable of direct interaction, the experiment did not address whether the interactions can occur within the cell. We therefore used the yeast two-hybrid assay and tested the domains of integrase and Atf1p for interactions. The two-hybrid assays detected the same interaction identified with the recombinant proteins, namely the binding of the b-ZIP domain of Atf1p to the catalytic core of integrase. The results suggested that integration was directed to the promoter of fbp1 by the binding of integrase to Atf1p anchored at UAS1. Working with recombinant proteins and DNA, we identified a three-component complex. Gel retardation assays detected a complex that contained integrase, the b-ZIP domain, and a 100 bp DNA from fbp1 that included UAS1. We also conducted experiments to test whether this complex was capable of directing integration. Integration products were detected within the 100 bp DNA that corresponded to the same positions of insertion that are selected in vivo. The data demonstrated that integration targeted to specific sites in the promoter of fbp1 was reconstituted with purified integrase, the b-ZIP domain of Atf1p, and a 100 bp DNA.
Ultra-high throughput sequencing of transposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe.
The finding that integration in the genome of S. pombe is directed to the promoters of genes raises several key questions about the biology of Tf1 integration, namely, whether all promoters are recognized equally or whether integration is directed to specific sets of promoters. If specific sets of promoters are preferred targets, what distinguishes the preferred promoters from those not recognized by Tf1. To address these questions, large numbers of integrations throughout the genome of S. pombe were sequenced (Guo and Levin, 2010). The revolutionary new methods for ultra high throughput sequencing made it possible to characterize extraordinarily large numbers of integration events.
To select for the cells with integration events, cells were induced for the expression of Tf1 containing neo (Tf1-neo). We applied ligation-mediated PCR to generate libraries of Tf1-neo associated with the downstream flanking DNA. We performed four independent transposition experiments (Hap_Mse_1, Hap_Mse_2, Dip_Mse, and Dip_Hpy), which were named according to the strains (haploid or diploid) and restriction enzymes (Mse I or Hpy CH4 IV) used to digest the genomic DNA from the cells with integration events. The cut libraries of DNA were ligated to linkers, and subjected to barcoded PCR. The amplified products, consisting of the downstream LTRs and their flanking DNA, were size selected and submitted to 454 Life Sciences for sequencing.
Altogether we obtained 599,760 high-quality sequence reads that were then analyzed with BLAST to determine the chromosomal location of the insertions. In all, there were 73,125 independent Tf1 integration events in unique positions of the S. pombe genome. The BLAST results of sequences from our first integration library identified 21,848 independent insertions in the experiment termed Hap_Mse_2. The insertions were broadly distributed across all three chromosomes. To examine the insertion data for preferences, all 21,848 insertion sites from the Hap_Mse_2 experiment were mapped relative to ORFs; the distance from the insertions to the closest ORF was determined. The integration from Hap_Mse_2 showed a clear preference for the first 500 nt upstream of ORFs.
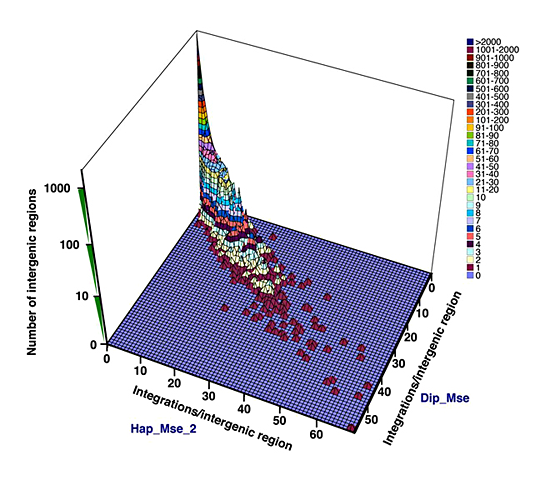
Figure 2. Comparison between two experiments (Hap_Mse_2 and Dip_Mse) of the number of insertions within each intergenic region
Each unit of the surface represents a group of intergenic regions. The X coordinate shows the number of integrations/intergenic region in the Hap_Mse_2 data. The Y coordinate shows the number of integrations/intergenic region in the Dip_Mse data. The Z coordinate has a log scale and shows the number of intergenic regions with the same X and Y coordinates. The colors of the surface and the associated key represent values of the Z coordinate.
The profile of integration across the genome revealed substantial variation, with some intervals containing 35 to 40 insertions per kb while many others had zero to five insertions per kb. An analysis of integration density for intervals of 10 kb also showed high levels of bias that were incompatible with random selection. The key question about this variation in integration is whether it could be attributed to intrinsic differences in integration efficiency between different sequences in the genome or whether the size of our cultures and the PCR amplification limited our ability to sample the integration potential of each sequence. To distinguish between these two possibilities, we tested whether the levels of integration in individual intergenic sequences were reproducible between multiple independent experiments. We compared the numbers of integration events in the intergenic regions of the Hap_Mse_2 experiment with the numbers of integration events from the Dip_Mse experiment (Figure 2). Each intergenic region was plotted using the number of integration events identified in the Hap_Mse_2 experiment as the X coordinate and the number of inserts recorded in Dip_Mse experiment as the Y coordinate. Because each of the 5,045 intergenic regions was plotted, and many intergenic regions had the same X,Y coordinates, we used the Z coordinate to indicate the number of the intergenic regions that had the same X,Y coordinates. The planar distribution of the data points shows that the level of integration in each intergenic region is similar between the two independent experiments. The R value for the data is 0.95 (R2=0.91), indicating that there is strong correlation of the integration levels between the two experiments. The comparison was performed between all pairs of the four experiments and the plots showed similar correlations.
In the Hap_Mse_2 experiment, 76% of all the insertion events occurred in just 20% of the intergenic sequences. The strong bias is a consequence of the integration preference for a specific set of promoters. One possibility was that Tf1 integrated into the promoters with the highest transcription activity. We tested this hypothesis but found no correlation between transcription and integration. In another effort to determine what distinguishes promoters that had high levels of insertions from the promoters that did not, we investigated whether the genes associated with the targeted promoters contributed to specific classes of biological function. The results of the gene ontology analysis suggested that genes regulated by environmental stress were among the strongest targets of integration. To examine this further, we sorted all the intergenic sequences from highest number of insertions to the lowest using the Hap_Mse_2 data. Using this order, the intergenic regions were placed into bins of 500 each. We then used published microarray data to tabulate how many of the intergenic regions in each bin contained promoters that are induced at least three-fold by conditions of stress. The bin containing the 500 intergenic regions with the most integration contained the highest number of genes induced by cadmium. The bins with successively lower amounts of integration contained fewer promoters that are induced by cadmium. This relationship indicates that integration has a preference for promoters that are induced by cadmium. Similar preferences were observed for genes induced when cells are treated with hydrogen peroxide or by heat. Particularly strong preferences for integration into promoters induced by methyl methanesulfonate or sorbitol were observed for the first bin of 500 intergenic regions. The targeting of Tf1 to stress-induced promoters represents a unique response that may function to specifically alter expression levels of stress response genes. Although there are no systematic data, integration of Tf1 into the promoter of ade6 and bub1 does stimulate transcription.
The size and number of the integration experiments reported here resulted in reproducible measures of integration for each intergenic region and ORF in the S. pombe genome. The reproducibility of the integration activity of each intergenic and ORF sequence from experiment to experiment demonstrates that we have saturated the full set of insertion sites that are actively targeted by Tf1. To our knowledge, this is the first time such a profile of integration data has been assembled.
Integration profiling: a whole-genome view of gene function
With the introduction of new deep sequencing technology, it is now possible to sequence many millions of transposon insertions in a single experiment. We tested whether Illumina sequencing could be used to generate a dense profile of transposon insertions that would reveal which genes are required for cell growth. For this experiment we used a haploid strain of S. pombe and Hermes, a DNA transposon from the housefly. In previous work we found that the Hermes transposon was highly active in S. pombe and that the insertions did not discriminate against ORFs. We predicted that, in actively growing cultures, Hermes insertions would not be tolerated in essential ORFs. This year, we induced Hermes transposition in a large S. pombe culture that was grown for 80 generations. With ligation-mediated PCR and Illumina sequencing, we were able to sequence 360,513 independent insertion events. On average, this represented one insertion for every 29 bp of the S. pombe genome. An analysis of integration density revealed that the ORFs largely separated into two classes, one with high numbers of insertions and another with much lower numbers. In collaboration with a group that deleted each of the genes of S. pombe, we found that the ORFs with low numbers of Hermes insertion corresponded to the essential genes. The ORFs with higher integration densities were in genes classified as nonessential. The results validated transposon profiling as a new method for identifying genes with essential function. Importantly, by applying specific conditions of selection during growth, this method can be adopted to identify genes that contribute to a wide variety of functions.
Additional Funding
- NIH Intramural AIDS Targeted Antiviral Program (2009 and 2010)
- Office of Intramural Research & Training AIDS Research Fellowship Award (2009)
Publications
- Park J, Evertts A, Levin H. The Hermes transposon of Musca domestica and its use as a mutagen of Schizosaccharomyces pombe. Methods. 2009; 49:243-247.
- Chatterjee AG, Leem Y, Kelly F, Levin H. The chromodomain of Tf1 integrase promotes binding to cDNA and mediates target site selection. J Virol. 2009; 83:2675-2685.
- Guo Y, Levin H.. High throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res. 2010; 20:239-248.
- Majumdar A, Chatterjee A, Ripmaster T, Levin H. The determinants that specify the integration pattern of retrotransposon Tf1 in the fbp1 promoter of Schizosaccharomyces pombe. J Virol. 2010; 29:In press.
Collaborators
- Shiv Grewel, PhD, Laboratory of Biochemistry and Molecular Biology, NCI, Bethesda, MD
Contact
For more information, email henry_levin@nih.gov or visit sete.nichd.nih.gov.


