You are here: Home > Section on Nutrient Control of Gene Expression
Transcriptional and Translational Regulatory Mechanisms in Nutrient Control of Gene Expression

- Alan G. Hinnebusch, PhD, Head, Section on Nutrient Control of Gene Expression
- Nazanin Ashourian, BS, Graduate Student
- Wen-Ling Chiu, PhD, Postdoctoral Fellow
- Kamal Dev, PhD, Postdoctoral Fellow
- Jinsheng Dong, PhD, Senior Research Assistant
- Naseem Gaur, PhD, Postdoctoral Fellow
- Daniel Ginsburg, PhD, Postdoctoral Fellow
- Cuihua Hu, BA, Special Volunteer
- Sebastien Lagiex, PhD, Postdoctoral Fellow
- Pilar Martin-Marcos, PhD, Postdoctoral Fellow
- Eun-Hee Park, PhD, Postdoctoral Fellow
- Hongfang Qiu, PhD, Staff Scientist
- Attiq Rehman, PhD, Postdoctoral Fellow
- Adesh Saini, PhD, Postdoctoral Fellow
- Fan Zhang, MS, Senior Research Assistant
- Fujun Zhou, PhD, Postdoctoral Fellow
We study the regulation of amino acid biosynthetic genes in budding yeast as a means of dissecting molecular mechanisms of gene regulation at the translational and transcriptional levels. Transcription of these and many other genes is coordinately induced by the transcriptional activator GCN4 in response to starvation for any amino acid. Expression of GCN4 is coupled to amino acid levels by a conserved translational control mechanism involving upstream open reading frames (uORFs) in GCN4 mRNA. Ribosomes translate the 5′-most uORF (uORF1), and under nonstarvation conditions, reinitiate translation at uORFs 2, 3 or 4 and then dissociate from the mRNA, keeping GCN4 translation repressed. In starvation conditions, the reinitiating ribosomes bypass uORFs 2–4 and reinitiate at GCN4 instead, owing to decreased availability of the ternary complex (TC)—comprised of initiation factor 2 (eIF2), GTP, and initiator Met-tRNAi—which binds to the small (40S) ribosomal subunit to assemble a 43S preinitiation complex (PIC). TC abundance is reduced in starved cells by phosphorylation of the alpha subunit of eIF2 (eIF2alpha) by Gcn2, a protein kinase conserved in all eukaryotes, converting eIF2 from substrate to inhibitor of its guanine nucleotide exchange factor (GEF) eIF2B. Hence, GCN4 translation is an in vivo indicator of impaired TC loading on 40S subunits. We previously exploited this to isolate mutations in subunits of eIF2B that constitutively derepress GCN4 (Gcd− phenotype) by lowering TC assembly in the absence of eIF2 phosphorylation. More recently, we used the Gcd− selection to identify domains/residues in eIF1, eIF1A, and eIF3, and also residues of 18S rRNA located near the "P" decoding site of the 40S subunit, that participate in rapid TC recruitment in vivo. In collaboration with Jon Lorsch's group, we demonstrated that segments/residues in eIF1, eIF1A, and 18S rRNA, which are implicated genetically in TC recruitment, also stimulate this reaction in a fully reconstituted in vitro system.
We also investigate the roles of various eIFs and the 40S subunit in scanning the mRNA 5′ untranslated region and accurately identifying the AUG initiation codon. The studies exploit a genetic selection for mutations that elevate initiation at near-cognate UUG start codons (Sui− phenotype) or suppress this aberrant initiation event (Ssu− phenotype). In this way, we provided the first evidence that eIF1A and the c-subunit of eIF3 make critical contributions to accurate AUG recognition, and we localized these functions to the unstructured N- and C-tails of eIF1A and the N-terminal domain of eIF3c. In collaboration with Lorsch's group, we also provided strong evidence that dissociation of eIF1 from the PIC is a critical step in AUG recognition, indicating that this factor serves as a "gate-keeper" necessary to suppress initiation at non-AUG triplets.
In the arena of transcriptional control, we previously identified the multiple co-activators required for gene activation by GCN4 and defined the molecular program for recruitment of nucleosome-remodeling enzymes and adaptor proteins that remove repressive chromatin structure and recruit TATA-binding protein, other general transcription factors, and RNA polymerase II (Pol II) to promoters. We also demonstrated the co-transcriptional recruitment of the co-activator SAGA, containing histone acetyltransferase (HAT) subunit Gcn5, to transcribed coding sequences and implicated SAGA/Gcn5 in transcription elongation.
Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome.
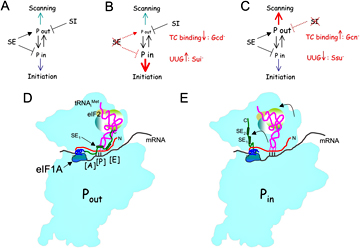
Figure 1. Proposed functions of eIF1A SE and SI elements in regulating different modes of TC binding to open and closed conformations of the 40S PIC
Structure of AP complexes and GGAs (A) Model describing the positive and negative effects of the SE and SI elements of eIF1A, respectively, on TC binding in the Pout conformation, which is conducive to scanning, and the second function of the SEs in blocking TC binding in the Pin conformation, which is incompatible with scanning and permissive for initiation. (B) SE inactivation destabilizes Pout, reducing TC loading and conferring the Gcd− phenotype, and also enhances the Pout-to-Pin transition at UUGs, conferring the Sui− phenotype. UUG initiation is further stimulated by loss of the inhibitory effect of the SEs on Pin. (C) SI inactivation stabilizes Pout, promoting TC loading and replacing Gcd- phenotypes with Gcn− phenotypes, and suppressing the Pout-to-Pin transition at UUGs to confer the Ssu− phenotype. (D-E) Hypothetical model depicting the different conformations of initiator tRNA in the Pin (D) and Pout (E) states and the proposed roles of the eIF1A SE elements in stabilizing initiator binding in Pout, where initiator is not fully accommodated in the P-site, and impeding initiator binding in Pin, where initiator is more fully engaged with the P-site. On AUG recognition, the SEs are ejected from the P-site to allow greater accommodation of initiator in the Pin state.
Recently, we discovered a 10-amino acid repeat in the C-terminal tail of eIF1A that is critical for both TC recruitment and accurate initiation at AUG start codons. Mutations in these repeats, dubbed scanning enhancers SE1 and SE2, were found to derepress GCN4 translation (Gcd− phenotype), signifying a reduced rate of TC loading on 40S subunits, which we confirmed in the reconstituted system in vitro.The SE mutations simultaneously increase initiation at UUG start codons in vivo, conferring the Sui− phenotype. Importantly, both the Sui− and Gcd− phenotypes, and the TC loading defect in vitro, conferred by SE mutations are suppressed by a substitution of residues 17-21 in the N-terminal segment of eIF1A, dubbed the scanning inhibitor (SI). These and other findings suggest that: 1) the eIF1A SEs promote TC binding to the 40S in a conformation compatible with scanning and the inspection of P-site triplets by Met-tRNAi but incompatible with start codon selection, which we dub POUT; and 2) block an alternative conformation incompatible with scanning but required for AUG recognition in which Met-tRNAi is fully accommodated in the P site, dubbed PIN.The SI element acts to antagonize the function of the SEs and destabilize POUT, thereby promoting PIN to enable start codon recognition when an AUG occupies the P-site (Figure 1A, D, E).Because TC initially binds to the 40S in the POUT conformation and subsequently isomerizes to the PIN state, our model explains why SE mutations, in destabilizing POUT, decrease the rate of TC loading but simultaneously allow the transition to PIN at UUG codons.By stabilizing POUT, SI mutations restore higher rates of TC loading and also suppress POUT to PIN transitions at UUG codons in SE mutants (Figure 1B-C). Our results provide strong evidence that eIF1A regulates start codon selection by controlling distinct modes of Met-tRNAi binding to the 40S P site (Saini et al., 2010).
The C-terminal region of eIF3a promotes mRNA recruitment, scanning and, together with eIF3j and the eIF3b RRM, selection of AUG start codons.
We previously presented evidence that eIF3, a 6-subunit complex that binds directly to the 40S subunit, functions in vivo to stimulate recruitment of TC in the assembly of 43S PICs and also attachment to mRNA to generate the 48S PIC. We also identified mutations in eIF3b or eIF3c that appear to impair scanning or alter the stringency of AUG selection. However, the molecular basis of these functions is not understood. In collaboration with Leos Valášek's group in Prague, we recently identified a discrete segment in the largest (a) subunit of eIF3, including the conserved "KERR" motif, that is required for attachment of the 43S PIC to mRNA, efficient scanning of long structured mRNA leaders, and efficient start codon recognition. Mutations in this segment that impair these functions also disrupt eIF3a binding to both the N-terminal segment of eIF3b and eIF3j, two subunits that bind to one another and also influence scanning and AUG recognition in vivo. Interestingly, the adjacent, extreme C-terminal region of eIF3a binds to the 40S proteins Rps2 and Rps3, which together with other published findings, locates the a-b-j module of eIF3 near the 40S mRNA entry channel. This location could enable the a-b-j module to regulate 43S attachment to mRNA, efficiency of scanning, and the transition between scanning-conducive and initiation-competent conformations of the PIC (Chiu et al., 2010).
The NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5.
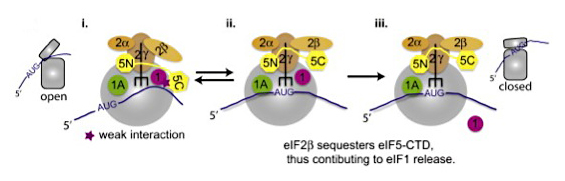
Figure 2. Model for two-stage co-transcriptional recruitment of histone deacetylase complex Rpd3C(S) to nucleosomes in coding regions
Structure of AP complexes and GGAs 1. Pol II is phosphorylated on Ser5 of the CTD by Kin28 during promoter clearance. Previous rounds of transcription leave ORF nucleosomes methylated on H3-K4 by Set1, more heavily towards the 5' end, and on H3-K36 by Set2, more heavily towards the 3' end. 2. Elongating Ser5-phosphorylated Pol II is phosphorylated on Ser2 by Bur1 and Ctk1, enabling efficient recruitment of Rpd3C(S) by direct interaction with Ser2-, Ser5-diphosphorylated CTD repeats. Subsequently, Rpd3C(S) is transferred from the phospho-CTD to H3 tails by interaction of H3-K36me2 with the chromodomain in Eaf3 and PHD finger in Rco1 (not depicted) for subsequent histone deacetylation.
We previously demonstrated that the HAT complex SAGA is co-transcriptionally recruited to coding sequences in a manner stimulated by phosphorylation of Serine 5 in the heptad repeats of the Pol II C-terminal domain (CTD) by the Cdk7/Kin28 subunit of TFIIH. We provided evidence that the HAT subunit (Gcn5) in SAGA acetylates histone H3 and facilitates transcription-coupled nucleosome eviction in coding sequences and that it also enhances the processivity of Pol II elongation and transcription-coupled histone H3-Lys4 methylation by Set1. Recently, we demonstrated that the histone H4 HAT complex NuA4 is also recruited co-transcriptionally to coding regions in a manner stimulated by Cdk7/Kin28 and that NuA4 association with nucleosomes further depends on H3 methylation, presumably due to chromodomains and a PHD finger in NuA4 subunits. We also obtained evidence that the HAT activities of NuA4 and SAGA cooperate to enhance co-transcriptional recruitment of the nucleosome remodeler RSC, promote histone eviction from transcribed sequences, and stimulate Pol II elongation rate. Consequently, inactivating Gcn5 and the HAT subunit in NuA4 (Esa1) confers additive reductions in transcript production from long versus short coding sequences in vivo. These findings demonstrate direct, additive roles for the HAT activities of NuA4 and SAGA in promoting the elongation phase of II transcription.
Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes.
We recently extended the two-stage recruitment mechanism, involving Serine5-phosphorylated Pol II CTD and methylated histone H3 described above for NuA4, to include the histone deacetylase complex (HDAC) Rpd3C(S).It was known that methylation of H3 by Set1 and Set2 is required for deacetylation of coding region nucleosomes by Set3C and Rpd3C(S), respectively. We discovered that Set3C and Rpd3C(S) are co-transcriptionally recruited to coding sequences in the absence of both Set1 and Set2, but in a manner stimulated by Cdk7/Kin28. We also showed that Rpd3C(S) and Set3C interact with both Pol II-Ser5P and histones in extracts; however, only the histone interactions require H3 methylation. Furthermore, a purified Rpd3C(S) complex, reconstituted from recombinant proteins by our collaborators Bing Li and Jerry Workman, was found to bind specifically to Ser5P synthetic peptides. Thus, whereas interaction with methylated H3 is required for Rpd3C(S) and Set3C deacetylation activities, their co-transcriptional recruitment is stimulated by the phosphorylated Pol II CTD. We further demonstrated that the HDAs Rpd3, Hos2, and Hda1 have overlapping functions in deacetylating nucleosomes and limiting co-transcriptional nucleosome eviction. Moreover, a strong correlation between increased acetylation and lower histone occupancy observed in single, double, and triple HDA mutants supports our contention, based on analysis of HAT mutants, that histone acetylation is a key determinant of co-transcriptional nucleosome eviction.
Activator GCN4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit Mediator to target genes in vivo.
The Mediator is a multisubunit coactivator required for transcription initiation by Pol II, which is recruited by Gcn4 to its target promoters in vivo.Previously, we determined that the tail subdomain of Mediator, containing the Gal11/Med15 subunit, is a direct target of Gcn4 in vivo, critical for recruitment by Gcn4 of both intact Mediator and the stable Mediator tail subdomain existing in sin4Δ cells. Although several Gal11 segments were shown previously to bind Gcn4 in vitro, the importance of these interactions for recruitment of Mediator and transcriptional activation by Gcn4 in cells was unknown. We demonstrated that interaction of Gcn4 with the Mediator tail in vitro and recruitment of this subcomplex and intact Mediator to the ARG1 promoter in vivo involve additive contributions from three different segments in the N-terminus of Gal11. These include the KIX domain, which is a critical target of other activators, and a region that shares a conserved motif (B-box) with mammalian coactivator SRC-1. We also established that the B-box is a critical determinant of Mediator recruitment by Gcn4. We further demonstrated that Gcn4 binds to the Gal11 KIX domain directly and, in collaboration with Christopher Jaroniec's group, utilized NMR chemical shift analysis, combined with mutational studies, to identify the likely Gcn4 binding site on the surface of the KIX domain. Together, the results define the physiological mechanism of Mediator recruitment by Gcn4. It appears that Gcn4 is distinctive in relying on comparable contributions from multiple segments of Gal11 for efficient recruitment of Mediator to target promoters in vivo.
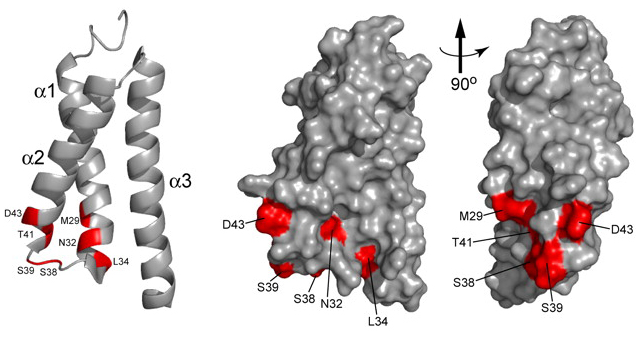
Figure 3. NMR analysis of interactions between the Med15/Gal11 KIX domain and the Gcn4 activation domain
Ribbon and surface representations of the Med15/Gal11 KIX domain with residues displaying amide chemical shift changes Δδ ≥ 0.10 ppm (highlighted in red) on interaction with the Gcn4 activation domain.
Additional Funding
- International Human Frontiers of Science Program Grant RGP0028
Publications
- Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol Cell Biol. 2009; 24:6473-6487.
- Chiu WL, Wagner S, Herrmannová A, Burela L, Zhang F, Saini AK, Valasek L, Hinnebusch AG. The C-terminal region of eIF3a promotes mRNA recruitment, scanning and, together with eIF3j and the eIF3b RRM, selection of AUG start codons. Mol Cell Biol. 2010; 30:4415-4434.
- Jedidi I, Zhang F, Qiu H, Stahl SJ, Palmer I, Kaufman JD, Nadaud PS, Mukherjee S, Wingfield PT, Jaroniec CP, Hinnebusch AG. Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit Mediator to target genes in vivo. J Biol Chem. 2010; 285:2438-2455.
- Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome. Genes Dev. 2010; 24::97-110.
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010; 39:234-246.
Collaborators
- Jo Lorsch, PhD, The Johns Hopkins University School of Medicine, Baltimore, MD
- Leos Valášek, PhD, Institute of Microbiology, Academy of Sciences of the Czech Republic, Prague, Czech Republic
- Paul Wingfield, PhD, National Institute of Arthritis and Musculoskeletal and Skin Diseases, NIH, Bethesda, MD
- Christopher Jaroniec, PhD, Ohio State University, Columbus, OH
- Bing Li, PhD, University of Texas Southwestern Medical Center at Dallas, Dallas, TX
- Jerry Workman, PhD, The Stowers Institute for Medical Research, Kansas City, MO
Contact
For more information, email hinnebua@mail.nih.gov or visit sncge.nichd.nih.gov.


