You are here: Home > Section on Macromolecular Recognition and Assembly
Intermolecular Forces, Recognition, and Dynamics

- Donald C. Rau, PhD, Head, Section on Macromolecular Recognition and Assembly
- Nina Yu Sidorova, PhD, Staff Scientist
- Rocco Ferrandino, BA, Postbaccalaureate Fellow
- Brandon Hoover, BA, Postbaccalaureate Fellow
- Benjamin Wiese, Special Volunteer
- Thomas Scott, Summer Intern
Our laboratory focuses on elucidating the coupling of the forces, structure, and dynamics of biologically important assemblies. The next challenge in structural biology is to understand the physics of interactions between molecules in aqueous solution. The ability to take exploit the increasing number of available protein and nucleic acid structures will depend critically on establishing the link between structures and binding energies. A fundamental and quantitative knowledge of intermolecular forces is necessary to understand the interactions among biologically important macromolecules that control cellular function and for rationally designing agents that can effectively compete with those interactions associated with disease. Our results have shown that experimentally measured forces at close spacings are similar for a wide variety of systems, but different from those predicted by current, conventionally accepted theories. We have interpreted the observed forces as indicating the dominating contribution from water structuring energetics (1). The observation that interacting macromolecules will tenaciously retain their hydration waters unless the surfaces are complementary has profound implications for recognition reactions. To investigate the role of water in binding, we measure and correlate changes in binding energies and hydration accompanying recognition reactions of biologically important macromolecules, particularly of sequence-specific DNA–protein complexes. We observe a strong correlation between retained water and binding energy; stronger binding means less water retained at the DNA–protein interface.
Direct force measurements
During vertebrate spermatogenesis, chromatin is dramatically reorganized by replacing histone with protamines to achieve a more compacted state for efficient genetic delivery and DNA protection. Given that DNA repair is inactive in sperm, dense packaging of DNA in sperm nuclei is considered important to protect DNA against damage by mutagens or oxidative species. Protamines are small, arginine-rich, nuclear proteins that condense the spermatid genome into an inactive state. Unlike histone-compacted DNA, the physical properties of reconstituted protamine–DNA assemblies closely resemble those of DNA condensed by multivalent cations that are much smaller and have much less charge. DNA is packaged by protamine to densities within the range seen for DNA condensed with the smaller multivalent ions. More importantly, the forces measured for reconstituted salmon protamine–DNA arrays by the osmotic stress technique coupled with x-ray diffraction show very similar characteristics to DNA precipitated by multivalent ions. We also showed that reconstituted salmon protamine DNA arrays and native salmon nuclei give the same osmotic stress force curves. We hypothesize that male infertility resulting from protamine deficiency and modification errors is attributable to looser packing of DNA in sperm nuclei than is optimal and hence greater accessibility to mutagens and oxidizing species. Mutations in sperm DNA have also been shown to correlate with birth defects.
Upon condensation with the smaller multivalent ions or protamines, compacted structures have well defined equilibrium separations of the DNA double helices of 0.7–1.5 nm, depending on the identity of the condensing ion. The finite separation of helices indicates a delicate balancing of a short-ranged repulsive force with a longer-ranged attraction. The physical origins of the forces acting to compact multivalent ion–DNA assemblies are still debated. We have argued for water-structuring or hydration forces on the basis of our previous extensive measurements of forces between both charged and uncharged molecules (1). Most theories of DNA assembly include some form of correlation of positive charges on one helix with negative phosphate charges on another in order to account for attraction. Using combined osmotic stress and single-molecule tweezing experiments, we previously characterized the distance dependence of the two component forces. Using this constraint, we can effectively separate the attractive and repulsive contributions of the force–distance curves.
Using synthetic peptides, we were able to elucidate several salient features of protamine compaction of DNA (2). All vertebrate protamines favor arginines almost exclusively over lysines. We found that arginine peptides can assemble DNA to much tighter spacings than lysine peptides of the same length. Packaging helices with protamines based on lysine in sperm nuclei would leave the DNA more vulnerable to damage. We hypothesize that differences in DNA–binding geometry between the two amino acids affect the correlation of charges on apposing helices. The incorporation of neutral amino acids into arginine peptides increases the amplitude of the short-ranged repulsive force while affecting attraction only slightly. We posit that the neutral amino acids increase the image charge–like repulsive force by displacing additional water from DNA grooves. The amplitudes of the repulsive and attractive forces for salmon protamine–DNA complexes can be quantitatively predicted from the number of arginines and the fraction of neutral amino acids. Incorporation of one negatively charged aspartate or phosphorylated serine into a hexa-arginine peptide increases the amplitude of the short-ranged repulsion dramatically—the equivalent of incorporating eight neutral amino acids. The longer-ranged attraction is moderately reduced but is consistent with a net +5 charge, with likely biological implications for spermatogenesis. The replacement of histones by protamines occurs in several steps. Serine phosphorylated protamines are precursors that initially bind to DNA. It is only after removing the phosphate groups that DNA is tightly packed. Incomplete dephosphorylation has been suggested a cause of increased DNA damage and male infertility. The results here show that phosphorylation has a much larger effect on weakening the net attraction between helices than simply reducing the protamine charge would suggest.
Click image to enlarge.
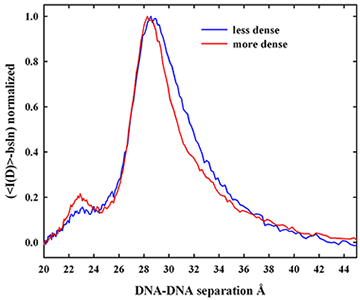
Figure 1. X-ray scattering from density gradient fractionated salmon sperm nuclei
The x-ray scattering intensity profiles are shown for density-fractionated salmon sperm nuclei. The more dense fraction has a narrower distribution of interhelical spacings with a peak spacing of about 29 Å. The two fractions overlap at the smaller distances, but the less dense fraction has a DNA population subset with significantly larger spacings. The difference between the two peaks has scattering peak at about 31.5 Å. The DNA with the larger spacings would be more accessible to mutagens and oxiding radicals.
We characterized the distribution of DNA–packing densities in native salmon sperm nuclei. Density gradient centrifugation is in standard use in assisted reproductive technologies (ART) and animal husbandry to obtain the best quality sperm for fertilization. We fractionated salmon sperm nuclei on a sucrose gradient and measured x-ray spacings between DNA helices. Figure 1 shows that more slowly sedimenting nuclei are characterized by an observably larger average spacing between helices than for denser fractions. Careful analysis of the scattering peaks indicates that the increased average spacing is the result of an increased spread of interhelical distances from normal to larger distances rather than a simple shift of a scattering peak with constant width to larger spacings. This may indicate a kinetic problem in forming protamine–DNA assembles.
There are several differences between mammalian and piscine protamines. The average fraction arginine of mammalian protamines, 50-55%, is significantly smaller than for fish, 65-70%. On the basis of our results, the increase in neutral amino acid content would be expected to decrease the net attraction, significantly increasing the equilibrium spacing between helices in mammalian sperm. A looser packaging of DNA owing to an increased fraction of neutral amino acids would increase accessibility of mutagens and reactive oxidizing species to the DNA, causing increased damage. This might explain another difference between mammalian and piscine protamine–DNA packaging. Extensive disulfide bridges between protamines are present only in mammalian sperm. The disulfide bridges in mammals may be required for tight DNA packaging, overcoming the increased fraction of neutral amino acids.
Many mammals have two protamines, P1 and P2, both longer than piscine protamines. We are now investigating DNA packaging in bull sperm. Bulls have only a P1 protamine and represent an intermediate in complexity between fish and humans sperm. The osmotic stress force curve for bull sperm nuclei with intact disulfide bonds is remarkably similar to that for salmon sperm nuclei. Reduction of the disulfide bonds with DTT leads to dramatic swelling of the nuclei (at least 10–20 fold in volume) and complete loss of x-ray order. The disulfide bonds are thus essential for dense packing of DNA. Once the sperm nucleus enters the egg, however, disulfide reduction results in significant unpacking of the DNA. Reducing agents as glutathione have been shown necessary for decondensation of mammalian sperm nuclei after fertilization. The osmotic stress force curve for DTT–reduced bull nuclei gradually merges at higher pressures with the unmodified bull sperm nuclei curve. Disulfide bonds do not reform if the DTT is removed from greatly swollen nuclei unless the nuclei are compacted with osmotic stress.
Future work includes the isolation of bull sperm protamine for reconstitution studies and density fractionation and x-ray analysis of bull sperm nuclei, coupled with determination of 8-oxoguanine by mass spectrometry as an indicator of oxidative DNA damage.
Hydration changes linked to sequence-specific DNA–protein recognition reactions
Our ultimate goal is to apply the lessons from direct force measurements to the recognition reactions that control cellular processes. We focus on differences in water sequestered by complexes of sequence specific DNA–binding proteins bound to different DNA sequences, with particular emphasis on correlating binding energy and water incorporated and on the energy necessary to remove hydrating water from complexes. We determine differences in sequestered water between complexes through the effect of changing osmotic pressure on binding constants or dissociation rates. Differences in water between specific and nonspecific complexes connect thermodynamics and structure. The emphasis on water allows us to approach recognition reactions in a direction that is different from standard practice. We take advantage of the unique experimental tools we developed to investigate the coupling of structure, thermodynamics, and function of the complexes from a fresh perspective.
The association and dissociation kinetics of sequence-specific DNA–binding proteins are surprisingly complicated. DNA–binding proteins that regulate gene activity locate their target sequences by initially binding nonspecifically, with subsequent one-dimensional diffusion along DNA interspersed with short hops or jumps and, it is thought, by direct transfer of protein from one DNA helix to another. Direct transfer is characterized by the formation of an intermediate DNA-protein-DNA ternary complex. Consequently, the dissociation rate depends on total DNA concentration. The direct transfer model is gaining popularity given that it is becoming more widely observed that protein–DNA dissociation rates depend on competitor DNA concentrations. Most specific and nonspecific complexes of these proteins with DNA entail charge-charge interactions, and thus the salt-concentration dependence of dissociation from direct transfer through a ternary DNA-protein complex should be much less than from simple dissociation. As the salt concentration is increased, the contribution from direct transfer to dissociation should significantly decrease. The difference, however, has not been directly investigated.
Click image to enlarge.
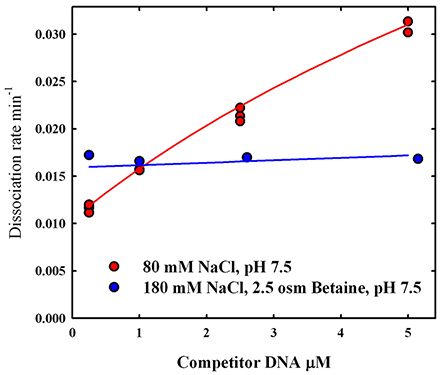
Figure 2. The dissociation rate dependence on competitor DNA concentration can be explained by kinetics of hopping.
The dissociation rate of EcoRI is shown as dependent on competitor DNA concentration for two salt concentrations. At 80 mM salt, the dependence on DNA concentration is commonly taken to indicate direct transfer from one DNA to another. The apparent finite intercept indicates an additional contribution from simple dissociation. At 180 mM salt, no DNA concentration dependence is observed. The solid lines are calculations of the dissociation rate for a model that explicitly includes protein 'hopping' excursions but not direct transfer.
We found that, at near physiological NaCl concentrations, the dissociation of the restriction endonuclease EcoRI from specific sequence DNA exhibits both competitor DNA concentration–dependent and –independent contributions (Figure 2). At two-fold higher salt concentrations, only the competitor oligonucleotide concentration–independent component is observed, consistent with the expected smaller salt-concentration dependence of direct transfer compared with simple dissociation. Unexpectedly, however, no difference is seen in the salt-concentration dependence of the dissociation rate measured around physiological salt concentration for two concentrations of competitor oligonucleotide that show quite different apparent contributions from direct transfer. These disparate observations cannot be explained by direct transfer, but can be rationalized by considering the physical consequences of transient protein dissociation. At short times after a protein dissociates from DNA, it remains in close proximity and has a high probability of rebinding to the same DNA. The probability that the protein will rebind after some time is given by the first passage time distribution function, which has been estimated for DNA–protein systems by simulations. During the time the protein is off the DNA, there is also a probability it will react with competitor DNA. This probability will depend on competitor DNA concentration and the association rate. Even if the protein should re-associate nonspecifically with the original DNA before reacting with competitor DNA, it could still dissociate before diffusing along the DNA far enough to encounter the recognition sequence. We previously characterized sliding distances for EcoRI as dependent on salt concentration (3). We developed an approximate model that incorporates a simulation-based first passage time distribution function, the sliding distance of EcoRI on DNA, and the binding kinetics to competitor DNA. As shown in Figure 2, the theory does fit the experimentally observed dependence of the dissociation rate on competitor concentration at physiological salt concentrations and the absence of competitor DNA concentration dependence at much higher salts. Since the protein has dissociated from the initial DNA and no ternary complex is formed, the salt dependence will be independent of oligonucleotide concentration. The difference in the salt concentration dependence at physiological salt concentration and at the higher salt concentration results from the difference in sliding distances. The protein can diffuse hundreds of base pairs along the DNA at physiological salt concentration. At much higher salt concentrations, however, the protein can only diffuse short distances along the DNA (tens of base pairs). It is highly unlikely that the protein can find the specific sequence before dissociating again. Reaction with the competitor DNA is much more probable now at all practical oligonucleotide concentrations.
A major biological role of restriction endonucleases is to protect bacteria from foreign DNA invasion. In the presence of Mg2+ ions, restriction endonucleases are precise molecular scissors, cleaving foreign, unmethylated DNA with exquisite specificity. The optimal enzymatic activity of EcoRV is reached at about pH 7.5. Cleavage activity decreases dramatically with decreasing pH while, paradoxically, specific equilibrium binding and binding specificity strongly increase (4). We found that, while specifically bound to its recognition site, the enzyme can bind to Mg2+and cleave DNA. We are now exploring the influence and interplay of pH, Mg2+ concentration, and osmotic stress on Mg2+ binding to the EcoRV–DNA complex, using a self-cleavage assay. The use of osmotic stress prevents significant dissociation and allows measurement Mg2+ binding and consequent cleavage on a convenient time scale. We find that the time dependence of cleavage after adding Mg2+ to EcoRV-DNA complexes exhibits a lag phase. The kinetics can be well fit considering sequential reactions with DNA cleavage occurring after the second step. The dependencies of each rate constant on solution variables are strikingly similar. Each is characterized by an uptake of about 30–40 water molecules, the binding of 1–2 Mg2+ ions, and the release of 1–2 H+ ions. We posit that the two steps correspond to the separate and independent binding of Mg2+ to each monomer of dimeric EcoRV. The osmotic pressure sensitivity is much less than for the specific-nonspecific transition, indicating that only a more restrained conformation change is necessary to bind Mg2+ in the catalytic site. We presume that the loss of enzymatic activity at lower pHs is attributable to titration of acidic residues in the catalytic site and consequent loss of Mg2+ binding affinity. The interplay of Mg2+ and pH might play an important role in the survival mechanism of enteric bacteria that spend a significant amount of time at very low pH conditions of the stomach.
Publications
- Stanley C, Rau DC. Evidence for water structuring forces between surfaces. Curr Opin Colloid Interface Sci 2011;16:551-556.
- DeRouchey JE, Rau DC. Role of amino acid insertions on intermolecular forces between arginine peptide condensed DNA helices: implications for protamine-DNA packaging in sperm. J Biol Chem 2011;286:41985-41992.
- Rau DC, Sidorova NY. Diffusion of the restriction nuclease EcoRI along DNA. J Mol Biol 2010;395:408-416.
- Sidorova NY, Muradymov S, Rau DC. Solution parameters modulating DNA binding specificity of the restriction endonuclease EcoRV. FEBS J 2011;278:2713-2727.
Collaborators
- Jason DeRouchey, PhD, University of Kentucky, Lexington, KY
- V. Adrian Parsegian, PhD, University of Massachusetts, Amherst, MA
- Xiangyun Qiu, PhD, George Washington University, Washington, DC
Contact
For more information, email raud@mail.nih.gov.

