You are here: Home > Section on Intracellular Protein Trafficking
Protein Sorting in the Endosomal-Lysosomal System

- Juan S. Bonifacino, PhD, Head, Section on Intracellular Protein Trafficking
- Rafael Mattera, PhD, Staff Scientist
- Xiaolin Zhu, RN, Technician
- Xuefeng Ren, PhD, Research Fellow
- Loreto Cuitiño, PhD, Visiting Fellow
- Ginny Farias, PhD, Visiting Fellow
- Xiaoli Guo, PhD, Visiting Fellow
- Shweta Jain, PhD, Visiting Fellow
- Sang-Yoon Park, PhD, Visiting Fellow
- Jing Pu, PhD, Visiting Fellow
- Christina A. Schindler, PhD, Visiting Fellow
- Stine C. Klinger, PhD, Guest Researcher
- Cecilia Bahamon, BS, Postbaccalaureate Student
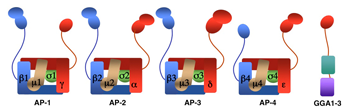
We investigate the molecular mechanisms by which transmembrane proteins are sorted to different compartments of the endo-membrane system, such as endosomes, lysosomes, lysosome-related organelles (e.g., melanosomes and platelet dense bodies), and specific domains of the plasma membrane in polarized cells (e.g., epithelial cells and neurons). Sorting is mediated by recognition of signals present in the cytosolic domains of transmembrane proteins by adaptor proteins that are components of membrane coats (e.g., clathrin coats). Among these adaptor proteins are the heterotetrameric AP-1, AP-2, AP-3, and AP-4 complexes (Figure 1), the monomeric GGA proteins (Figure 1), and the heteropentameric retromer complex. Proper sorting requires the function of additional components of the trafficking machinery that mediate vesicle tethering and fusion, such as the multisubunit tethering complex GARP. Current work in our laboratory is aimed at elucidating the structure, regulation, and physiological roles of coat proteins and vesicle-tethering factors, as well as investigating human diseases that result from genetic defects of these proteins (e.g., Hermansky-Pudlak syndrome; neurodegenerative and neurodevelopmental disorders) or their subversion by human pathogens (e.g., HIV-1).
An AP-1/clathrin pathway for the sorting of transmembrane receptors to the somatodendritic domain of hippocampal neurons
Click image to enlarge.
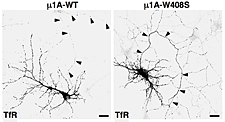
Figure 2. Overexpression of dominant-negative mutant of AP-1 subunit (μ1A-W408S) causes missorting of transferrin receptor (TfR) to the axon (arrowheads).
AP-1, AP-2, and AP-3 are clathrin-associated adaptor complexes that recognize two types of sorting signal, referred to as tyrosine-based and dileucine-based signals. Previous studies from our laboratory showed that tyrosine-based signals bind to the mu1, mu2, and mu3 subunits, whereas dileucine-based signals bind to a combination (i.e., a hemicomplex) of two subunits—gamma-sigma1, alpha-sigma2, and delta-sigma3—from the corresponding AP complexes. A major goal of our recent work is the analysis of the role of signal-adaptor interactions in polarized sorting in neurons. Neurons are polarized into dendrites, soma, and axons. The plasma membrane of each of these domains possesses a distinct set of transmembrane proteins, including receptors, channels, transporters, and adhesion molecules. We hypothesized that sorting to these domains is mediated by interaction of sorting signals with AP complexes. Our studies showed that the cytosolic tails of various transmembrane receptors, including the transferrin receptor (TfR), the Coxsackie virus and adenovirus receptor (CAR), the Nipah virus F glycoprotein (NiV-F), and the glutamate receptor proteins mGluR1, NR2A, and NR2B, all have information leading to the sorting of these proteins to the somatodendritic domain of hippocampal neurons. In the case of TfR, CAR, and NiV-F, this information occurs in the form of tyrosine-based sorting signals. Protein interaction analyses showed that the tails of these receptors bind to the mu1A subunit of AP-1. Dominant-negative interference and RNAi approaches demonstrated that the interaction of cytosolic tails with AP-1 is responsible for somatodendritic sorting (Figure 2). Sorting involves exclusion of the receptor proteins from vesicular transport carriers destined for the axonal domain at the level of the soma. Interference with AP-1–dependent somatodendritic sorting caused defective maturation of dendritic spines and reduced the number of excitatory synapses.
An AP-1/clathrin pathway for the sorting of transmembrane receptors to the somatodendritic domain of hippocampal neurons
A major effort of our laboratory this past year was to examine the role of signal-adaptor interactions in polarized sorting in neurons (2). Neurons are polarized into dendrites, soma, and axons. The plasma membrane of each of these domains possesses a distinct set of transmembrane proteins, including receptors, channels, transporters, and adhesion molecules. We hypothesized that sorting to these domains could be mediated by interaction of sorting signals with AP complexes. Our studies showed that the cytosolic tails of various transmembrane receptors, including the transferrin receptor (TfR), the Coxsackie virus and adenovirus receptor (CAR), and the glutamate receptor proteins mGluR1, NR2A, and NR2B, all have information leading to their sorting to the somatodendritic domain of hippocampal neurons. In the case of TfR and CAR, this information exists in the form of tyrosine-based sorting signals. Protein interaction analyses showed that the tails of these receptors bind to the μ1A subunit of AP-1. Dominant-negative interference and RNAi approaches demonstrated that interaction of cytosolic tails with AP-1 was responsible for somatodendritic sorting. For example, overexpression of a dominant-negative mutant of the μ1A subunit of AP-1 (μ1A-W408S), but not of the wild-type protein (μ1A-WT), resulted in the appearance of transferrin receptor (TfR) in the axon (Figure 2). Sorting involved exclusion of the receptor proteins from transport carriers destined for the axonal domain at the level of the soma. Interference with AP-1–dependent somatodendritic sorting caused defective maturation of dendritic spines and reduced the number of synapses. Recently, mutations in the sigma1A and sigma1B subunits of AP-1 were shown to be the cause of two neurodevelopmental disorders known as MEDNIK syndrome and Fried syndrome, respectively. Our findings suggest that these disorders may arise from failure to sort certain cargoes to the somatodendritic domain of specific neuronal populations.
Recently, mutations in the sigma1A and sigma1B subunits of AP-1 were shown to be the cause of two syndromic mental retardation disorders, known as MEDNIK syndrome and Fried syndrome, respectively. Unlike mu1A, sigma1A and sigma1B recognize dileucine-based sorting signals. Our findings suggest that MEDNIK and Fried syndromes arise from failure to sort dileucine-containing cargos to the somatodendritic domain of specific neuronal populations. Together with previous findings in epithelial cells, our recent results establish AP-1 as a global regulator of polarized sorting in various cell types.
Structural bases for membrane recruitment and activation of the AP-1 adaptor complex
Structural studies provided valuable insights into the mechanisms by which AP complexes recognize sorting signals. In collaboration with James Hurley, we elucidated the mechanism by which the small GTPase Arf1 recruits the AP-1 complex to membranes and induces a conformational change that allows AP-1 to bind to both tyrosine-based and dileucine-based sorting signals. X-ray crystallographic analysis of Arf1 in complex with the AP-1 core showed that two molecules of Arf1-GTP bind to distinct sites on the gamma and beta1 subunits of AP-1. The AP-1 core in this complex is in the open conformation, which exposes binding sites for tyrosine-based and dileucine-based sorting signals. The structure reveals how Arf1-induced conformational activation couples membrane recruitment to sorting-signal recognition.
Mechanisms of signal recognition by the AP-3 complex
In addition to AP-1, we also studied the mechanisms of signal recognition by the related AP-3 complex. In collaboration with the group of Michael Marks, we showed that recognition of a dileucine-based sorting signal in the cytosolic tail of the oculocutaneous albinism type 2 (OCA2) protein by both AP-1 and AP-3 mediates OCA2 sorting to melanosomes. In a separate study, we solved the crystal structure of the mu3A subunit of AP-3 in complex with a tyrosine-based sorting signal. The structure revealed that the binding site on mu3A is similar to that on the mu2 subunit of AP-2. These studies contributed to the elucidation of the mechanisms by which the AP-3 complex sorts transmembrane proteins to melanosomes, thus shedding light on the pathogenesis of the pigmentation and bleeding disorder Hermansky-Pudlak syndrome type 2.
Structural insights into the mechanisms of vesicle tethering and fusion
In collaboration with Aitor Hierro, we uncovered the structural mechanism by which the tethering factor GARP interacts with its cognate vesicle fusion SNARE proteins. X-ray crystallographic analyses showed how the Ang2 subunit of GARP binds to the Habc domain of the Syntaxin 6 SNARE. Our findings highlight a key event in the pathway by which transport vesicles emanating from endosomes fuse with the trans-Golgi network. The integrity of this pathway is essential for neuronal function and viability, and its perturbation leads to neurodegenerative movement disorders.
Mechanisms of CD4 downregulation by the Vpu protein of HIV-1
Primate immunodeficiency viruses target helper T-cells and macrophages/monocytes through binding of the viral envelope glycoprotein to a combination of CD4 and a chemokine receptor (CCR4 or CXCR5) on the surface of host cells. Strikingly, infection results in rapid and sustained downregulation of CD4 and, to a lesser extent, of the chemokine receptors. Downregulation of these viral co-receptors prevents superinfection, promotes virion release, and interferes with the immune response, leading to the establishment of a robust infection. CD4 downregulation is so important to the life cycle of human immunodeficiency virus-1 (HIV-1) that two accessory proteins, Nef and Vpu, encoded in the viral genome, are devoted to this task. Indeed, Nef and Vpu are critical for the progression from infection to AIDS, a fact that is best illustrated by the existence of long-term non-progressors who are infected with HIV-1 strains bearing inactivating mutations in the genes encoding these proteins. Therefore, pharmacologic or biologic perturbation of Nef and/or Vpu has the potential to prevent the pathogenic effects of HIV-1. To date, however, this potential has not been realized, mainly because Nef and Vpu have no enzymatic activity and their mechanisms of action are insufficiently understood.
In previous work, we made substantial progress towards elucidating the mechanism of CD4 downregulation by Nef. We found that Nef connects surface CD4 to both the endocytic and lysosomal targeting machineries, leading to efficient and sustained removal of CD4 from host cells early during infection. The current project focuses on the mechanisms by which Vpu downregulates CD4 at later stages of infection. Vpu is a small transmembrane protein comprising a short luminal domain, a single transmembrane domain (TMD), and a cytosolic domain. The Vpu cytosolic domain binds simultaneously to the CD4 cytosolic tail and to the SCF-beta-TrCP E3 ubiquitin ligase complex in the endoplasmic reticulum (ER), causing CD4 ubiquitination on lysine and serine/threonine residues, followed by interaction with the VCP-UFD1L-NPL4 dislocase complex and subsequent targeting for degradation by the proteasome. Recognition of CD4 for Vpu-induced degradation involves interactions at the level of both the cytosolic and transmembrane domains of CD4 and Vpu. Transmembrane domain interactions promote targeting for degradation as well as retention of CD4 in the ER. Interference with transmembrane domain interactions reduces CD4 polyubiquitination as well as dislocation from the ER membrane, thus abrogating CD4 degradation in vivo. The multiple levels at which Vpu engages the cellular quality control mechanisms underscore the importance of ensuring profound suppression of CD4 for the life cycle of HIV-1.
In collaboration with Ramanujan Hegde, we succeeded this past year in reconstituting Vpu-induced CD4 ubiquitination in an in vitro system. Also in this system, CD4 and Vpu engaged in transmembrane domain interactions. Disruption of these interactions reduced the extent of CD4 polyubiquitination. Unexpectedly, differential processivity alone could not explain the differing fates of degraded and nondegraded transmembrane CD4 variants. Instead, we identified continuous deubiquitination as a prerequisite for maximal substrate discrimination. These findings explain how small differences in substrate-ligase interaction can be amplified into larger differences in polyubiquitination and net degradation. In addition to shedding light on the mechanisms of Vpu-induced CD4 degradation, our results provide a conceptual framework for the general mechanisms of substrate discrimination during membrane protein quality control.
Additional Funding
- Intramural AIDS Targeted Antiviral Program (IATAP)
Publications
- Farías GG, Cuitino L, Guo X, Ren X, Jarnik M, Mattera R, Bonifacino JS. Signal-mediated, AP-1/clathrin-dependent sorting of transmembrane receptors to the somatodendritic domain of hippocampal neurons. Neuron 2012;75:810-823.
- Ren X, Farías GG, Canagarajah BJ, Bonifacino JS, Hurley JH. Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell 2013;152:755-767.
- Mardones GA, Burgos PV, Lin Y, Kloer DP, Magadán JG, Hurley JH, Bonifacino JS. Structural basis for the recognition of tyrosine-based sorting signals by the mu3A subunit of the AP-3 adaptor complex. J Biol Chem 2013;288:9563-9571.
- Zhang ZR, Bonifacino JS, Hegde R. Deubiquitinases sharpen substrate discrimination during membrane protein degradation from the ER. Cell 2013;164:609-622.
- Abascal-Palacios G, Schindler C, Rojas A, Bonifacino JS, Hierro A. Structural basis for the interaction of the Golgi-Associated Retrograde Protein (GARP) complex with the t-SNARE Syntaxin 6. Structure 2013;21:1698-1706.
Collaborators
- Ramanujan Hegde, MD, PhD, MRC Laboratory of Molecular Biology, Cambridge, UK
- Aitor Hierro, PhD, CIC-bioGUNE, Bilbao, Spain
- James Hurley, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
- Michael S. Marks, PhD, University of Pennsylvania School of Medicine, Philadelphia, PA
Contact
For further information, contact juan@helix.nih.gov or visit cbmp.nichd.nih.gov/sipt.