You are here: Home > Unit on Microbial Pathogenesis
Host Cell Modulation by the Intracellular Pathogen Legionella pneumophila

- Matthias Machner, PhD, Head, Unit on Microbial Pathogenesis
- Kimberly Decker, PhD, Postdoctoral Fellow
- Andrew Gaspar, PhD, Postdoctoral Fellow
- Yi-han Lin, PhD, Postdoctoral Fellow
- M. Ramona Neunuebel, PhD, Postdoctoral Fellow
We investigate the molecular mechanisms that allow microbial pathogens to exploit the human host and cause disease. As a model system we use the bacterium Legionella pneumophila, the causative agent of a potentially life-threatening pneumonia called Legionnaires' disease. Upon phagocytosis by alveolar macrophages, L. pneumophila delivers a large number of bacterial proteins, or effectors, into the host cell cytosol through the Dot/Icm type IV secretion system (T4SS) (Figure 1). Some effectors help L. pneumophila redirect proteins and membrane material of the infected cell to its Legionella-containing vacuole (LCV), thereby establishing a protective compartment resembling host-cell endoplasmic reticulum (ER). L. pneumophila mutants with a non-functional T4SS are avirulent, underscoring the importance of the translocated effector proteins for L. pneumophila infection. The main focus of our laboratory is to identify and characterize host–pathogen interactions during L. pneumophila infection and to determine their importance for bacterial virulence.
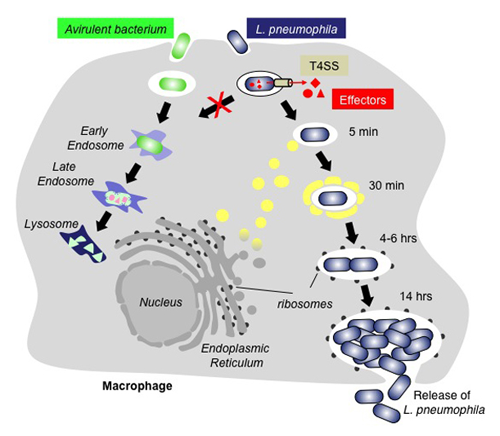
Figure 1. The infection cycle of L. pneumophila
Intracellular trafficking of Legionella pneumophila (blue) or an avirulent bacterium (green). Upon uptake by a macrophage cell, the phagosome containing an avirulent bacterium traffics along the endosomal route and gradually matures into a lysosome, which kills and degrades the bacterium. In contrast, L. pneumophila can escape the default endo-lysosomal pathway with the help of its effector proteins (red) that enter the host cell through the Dot/Icm type IV secretion system (T4SS; beige). Proteins and transport vesicles (yellow spheres) are recruited to the Legionella-containing vacuole (LCV), which is slowly transformed into a ribosome-studded compartment that mimics host-cell rough endoplasmic reticulum (ER) and supports replication of L. pneumophila. Eventually, the host cell is lyzed, and L. pneumophila bacteria are released to infect other macrophages.
Rab1 dynamics on LCVs is regulated by multiple L. pneumophila effector proteins.
L. pneumophila intercepts transport vesicles from the early secretory pathway between the endoplasmic reticulum (ER) and the Golgi compartment. Rab1, a key regulator of this pathway, transiently localizes to LCVs (Figure 2), suggesting that L. pneumophila utilizes Rab1 in order to redirect secretory vesicle traffic to its vacuolar membrane. We recently discovered that the effector protein SidM is critical for Rab1 exploitation. SidM combines the activity of a GDP–dissociation inhibitor (GDI) displacement factor (GDF) with a guanine nucleotide exchange factor (GEF). SidM mediates dissociation of Rab1 from its chaperone GDI and Rab1 activation by catalyzing exchange of GDP for GTP in Rab1. Bacterial mutants lacking SidM are defective for Rab1 recruitment, indicating that SidM is essential to exploit the pool of GDI–bound Rab1 during infection. SidM is the first known example of a protein shown to combine GDI displacement and nucleotide exchange activity.
In addition to its GEF/GDF activity, SidM also catalyzes the covalent attachment of adenosine monophosphate (AMP) to tyrosine-77 of Rab1, a process known as AMPylation. AMPylated Rab1 is locked in the active conformation because it cannot be inactivated by GTPase–activating proteins (GAPs), including the L. pneumophila effector LepB. AMPylated Rab1 is believed to attract cellular cargo vesicles to the LCV without interference from host proteins that would otherwise deactivate Rab1.
We recently discovered that, in addition to encoding the AMPylase SidM, L. pneumophila produces the effector protein SidD, which catalyzes AMP removal from Rab1, a process known as de-AMPylation (or de-adenylylation). Using purified proteins in in-vitro de-AMPylation studies, we found that, once AMP is removed from Rab1 by SidD, de-AMPylated Rab1 is accessible for inactivation by GAP proteins such as L. pneumophila LepB. Using indirect immunofluorescence microscopy, we found that L. pneumophila mutants lacking SidD showed a prolonged colocalization with host-cell Rab1, demonstrating that SidD is required for the timely removal of Rab1 from LCVs. The identification of L. pneumophila SidD as Rab1 de-AMPylase not only provides the missing link in a complex chain of Rab1 modulation events during L. pneumophila infection but also represents an intriguing example for how prokaryotic and possibly eukaryotic cells may regulate protein function through reversible AMPylation.
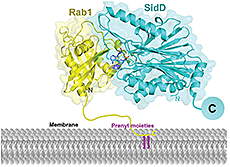
In collaboration with the laboratory of Aitor Hierro, we also succeeded in solving the crystal structure of the catalytic domain of SidD (Figure 3). The structure reveals a phosphatase-like fold with additional structural elements not present in generic PP2C-type phosphatases. The active side contains a binuclear metal-binding site characteristic of hydrolytic metalloenzymes, with strong dependency on magnesium ions. Subsequent docking and molecular dynamics simulations between SidD and Rab1 revealed the interface contacts and the energetic contribution of key residues to the interaction. Given that the de-AMPylation reaction is chemically identical to a de-phosphorylation reaction, our findings suggest that L. pneumophila SidD is a converted phosphatase designed to accept the larger AMP side chain as substrate.
Exploitation of host cell Rab6A′ by the L. pneumophila effector protein LidA
Click image to enlarge.
Figure 3. Structural model of the interaction between SidD and Rab1 on the vacuolar membrane
This ribbon diagram represents the structure of the complex between SidD (cyan) and AMPylated Rab1 (yellow) from the last nanosecond of a molecular dynamics simulation. Metal ions are shown as green spheres, and the AMPylated tyrosine side chain of Rab1 is shown as stick model.
While the function of SidM, SidD, and LepB has been characterized in great detail, the role of LidA, another Rab1– and Rab6A′–interacting effector protein from L. pneumophila, is poorly understood. We found that LidA preferentially binds to Rab6A′ in the active GTP–bound conformation. Rab6 binding occurrs through the central region of LidA and followed a stoichiometry of 1:2 for the LidA–Rab6A′ complex. LidA maintains Rab6A′ in the active conformation by efficiently blocking the hydrolysis of GTP by Rab6A′, even in the presence of cellular GTPase–activating proteins, suggesting that the function of Rab6A′ must be important for efficient intracellular replication of L. pneumophila. Accordingly, we found that production of constitutively inactive Rab6A′(T27N) but not constitutively active Rab6A′(Q72L) significantly reduces the ability of L. pneumophila to initiate intracellular replication in human macrophages. Thus, the presence of an active pool of Rab6 within host cells early during infection is required to support efficient intracellular growth of L. pneumophila.
Additional Funding
- NICHD Director's Challenge Innovation Award
Publications
- Chen Y, Tascón I, Neunuebel MR, Pallara C, Brady J, Kinch LN, Fernández-Recio J, Rojas AL, Machner MP, Hierro A. Structural basis for Rab1 de-AMPylation by the Legionella pneumophila effector SidD. PLoS Pathogens 2013;9:e1003382.
- Chen Y, Machner MP. Targeting of the small GTPase Rab6A’ by the Legionella pneumophila effector LidA. Infect Immun 2013;81:2226-2235.
- Neunuebel MR, Mohammadi S, Jarnik M, Machner MP. Legionella pneumophila LidA affects nucleotide binding and activity of the host GTPase Rab1. J Bacteriol 2012;194:1389-1400.
Collaborators
- Aitor Hierro, PhD, CIC bioGUNE Institute, Bilbao, Spain
- Michal Jarnik, PhD, Cell Biology and Metabolism Program, NICHD, Bethesda, MD
Contact
For more information, email machnerm@mail.nih.gov or visit cbmp.nichd.nih.gov/ump/research.html