You are here: Home > Section on Organelle Biology
Interplay between Membrane Organelles, Cytoskeleton and Metabolism in Cell Organization and Function

- Jennifer Lippincott-Schwartz, PhD, Head, Section on Organelle Biology
- Yu Chen, PhD, Visiting Fellow
- Sarah Cohen, PhD, Visiting Fellow
- Dylan Burnette, PhD, Postdoctoral Fellow
- Antony Chen, PhD, Postdoctoral Fellow
- CJ Hsu, PhD, Postdoctoral Fellow
- Uri Manor, PhD, Postdoctoral Fellow
- Carolyn Ott, PhD, Postdoctoral Fellow
- Angelika Rambold, PhD, Postdoctoral Fellow
- Prabuddha Sengupta, PhD, Postdoctoral Fellow
- Arnold Seo, PhD, Postdoctoral Fellow
- Alex Valm, PhD, Postdoctoral Fellow
- Schuyler Van Engelenburg, PhD, Postdoctoral Fellow
- Pick Wei, PhD, Postdoctoral Fellow
- Prasanna Satpute, PhD, Volunteer
- Alex Ritter, BA, Graduate Student
- Bennett Waxse, BA, Graduate student
We investigate the global principles underlying cell behavior at both small and large spatial scales. At the small scale, we employ the super-resolution imaging techniques of photoactivated localization microscopy (PALM), interferometric 3D PALM, single-particle tracking PALM, and pair-correlation PALM to map the spatial organization, stoichiometry, and dynamics of proteins associated with various membrane-bound compartments and with the cytoskeleton. We also employ fluorescence photobleaching, photoactivation, fluorescence correlation, and fluorescence energy transfer methods to measure protein-protein interactions, protein turnover rates, and protein association rates. Such approaches allow us to assay cellular functions, including receptor stoichiometry and protein clustering and diffusion behavior at the nanometric scale in living cells. At the large scale, we investigate how complex behaviors of cells arise, such as cell crawling (Figure 1), polarization, furrowing, cytokinesis, cell fate determination, viral budding, and intercellular transfer. We study these intricate behaviors by quantitatively analyzing diverse intracellular processes, including membrane trafficking, autophagy, actin/microtubule dynamics, and organelle assembly/disassembly pathways, which undergo dramatic changes as cells alter their behavior and organization throughout life. To assist with these efforts, we combine various fluorescence-based imaging approaches, including TIRF imaging and spinning-disk and laser-scanning confocal microscopy, with FRAP, FLIP, and photoactivation to obtain large image data sets. We process the data sets computationally to extract biochemical and biophysical parameters, which can be related to results from conventional biochemical assays. We then use the results to generate mechanistic understanding and predictive models of the behavior of cells and subcellular structures (including ER, Golgi, cilia, endosomes, lysosomes, autophagosomes, and mitochondria [Figure 2]) under healthy and pathological conditions.
Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes.
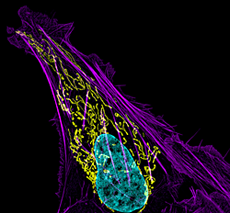
Click image to enlarge.
Figure 1. Mitochondral and actin organization in a crawling cell
SIM image of MITO-RFP, Phalloidin-Alexa 488, and DNA (Hoeschst 33342) in a U2OS cell (osteoblastoma)
Rab proteins are important regulators of insulin-stimulated GLUT4 translocation to the plasma membrane (PM), but the precise steps in GLUT4 trafficking modulated by particular Rab proteins remain unclear. To clarify the issue, we used TIRF microscopy to monitor GLUT4 trafficking under basal conditions and during insulin signaling, identifying Rab proteins associated with GLUT4 storage vesicles (GSVs) and their mode of action on GLUT4 trafficking. Screening a large set of Rab proteins for colocalization with GLUT4, for association with IRAP–pHluorin vesicles fusing at the PM in response to insulin stimulation, and for PM recruitment under insulin stimulation, we discovered that only Rab10 associates with GSVs and mediates GSV peripheral translocation after becoming activated. The Rab GTPase activation protein AS160 negatively regulated Rab10 activity on GSVs. Furthermore, we found that Rab10 interacted with the actin motor protein myosin-Va, which facilitated the translocation of GSVs to sites at the PM where the vesicles could fuse. High expression levels of a short tail form of myosin-Va lacking the actin-binding domain reduced GLUT4 delivery to the PM. Thus, even though multiple Rab proteins regulate the trafficking of GLUT4, it is only Rab10 in coordination with myosin-Va that mediates the final steps of GSV translocation to the PM under insulin stimulation.
Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling
The physical separation of two daughter cells at the end of mitosis, known as cytokinetic abscission, involves cleavage of a narrow, microtubule-based, intercellular bridge that connects two nascent daughter cells arising during cell division. We have been investigating the role of the endosomal sorting complex required for transport (ESCRT)-III complex in this process. Using high-resolution, quantitative imaging of ESCRT-III during cytokinetic abscission, we observed that ESCRT-III initially assembles at the midbody dark zone and then polymerizes outward to the site of cytokinetic abscission. Integrating the observations with the known biophysical properties of ESCRT-III complexes, we formulated and tested a computational model for ESCRT–mediated cytokinetic abscission. In the model, ESCRT-III forms a fission complex that drives constriction and abscission of the intercellular cytokinetic bridge. The ESCRT-III fission complex arises as a result of breakage enabled by the ATPase VPS4 of the initial ESCRT-III oligomer, polymerizing at the edge of the midbody dark zone. Once formed, the fission complex contracts to its spontaneous diameter of about 50 nm, while sliding along the intercellular bridge away from the midbody dark zone. Sliding continues until the fission complex reaches its equilibrium position corresponding to the minimal elastic energy of the bridge membrane. The subsequent abscission is driven by attachment of the bridge membrane to the dome-like end-cap of the fission complex. We substantiated this model by theoretical analysis of the membrane elastic energy and by experimental verification of the major model assumptions. Currently, we are exploring whether ESCRT-III polymerization coupled with breakage and sliding of a constricting membrane-bound ESCRT-III complex to a location corresponding to minimal elastic energy of the membrane is a conserved property of other ESCRT–mediated processes (i.e., human immunodeficiency virus budding).
Evidence for a metabolic shift during cell polarization

Click image to enlarge.
Figure 2. Mitochondria morphology in NRK cells
Image, obtained by structured illumination microscopy, of mitochondria labeled with GFP-tagged prohibitin
Cell polarization requires increased cellular energy and metabolic output, but how these energetic demands are met by polarizing cells is unclear. To address these issues, we investigated the roles of mitochondrial bioenergetics and autophagy during cell polarization of hepatocytes cultured in a collagen sandwich system. We found that, as the hepatocytes begin to polarize, they use oxidative phosphorylation to raise their ATP levels and that the energy production is required for polarization. After the cells are polarized, the hepatocytes shift to become more dependent on glycolysis to produce ATP. Along with this central reliance on oxidative phosphorylation as the main source of ATP production in polarizing cultures, other metabolic processes are reprogrammed during the course of polarization. As the cells polarize, mitochondria elongate, and mitochondrial membrane potential increases. In addition, lipid droplet abundance falls over time. The findings suggest that polarizing cells are reliant on fatty acid oxidation, which is supported by pharmacologic inhibition of β-oxidation by etomoxir. Finally, autophagy is up-regulated during cell polarization, with inhibition of autophagy retarding cell polarization. Taken together, our results describe a metabolic shift involving several coordinated metabolic pathways that ultimately serve to increase energy production during cell polarization.
Fast structural responses of gap junction membrane domains to AB5 toxins
Gap junctions (GJs) represent connexin-rich membrane domains that connect interiors of adjoining cells in mammalian tissues. How fast GJs can respond to bacterial pathogens was not previously known. Using Bessel beam plane illumination and confocal spinning disk microscopy, we found fast (about 500 ms) formation of connexin-depleted regions (CDRs) inside GJ plaques between cells exposed to AB5 toxins. CDR formation appears as a fast redistribution of connexin channels within GJ plaques with minor changes in outline or geometry. CDR formation does not depend on membrane trafficking or the submembrane cytoskeleton and has no effect on GJ conductance. However, CDR responses depend on membrane lipids, can be modified by cholesterol-clustering agents and extracellular K+ ion concentration, and influence cAMP signaling. The CDR response of GJ plaques to bacterial toxins is a phenomenon observed for all tested connexin isoforms. Through signaling, the CDR response may enable cells to sense exposure to AB5 toxins. CDR formation may reflect lipid-phase separation events in the biological membrane of the GJ plaque, leading to increased connexin packing and lipid reorganization. Our data demonstrate very fast dynamics (in the millisecond-to-second range) within GJ plaques, which previously were considered to be relatively stable, long-lived structures.
Insulin triggers direct trafficking to the cell surface of sequestered GLUT4 storage vesicles marked by Rab10.
Understanding how the glucose transporter isoform 4 (GLUT4) redistributes to the plasma membrane during insulin stimulation is a major goal of glucose transporter research. GLUT4 molecules normally reside in numerous intracellular compartments, including specialized storage vesicles and early/late endosomes. It is unclear how these diverse compartments respond to insulin stimulation to deliver GLUT4 molecules to the plasma membrane. For example, we do not know whether they fuse with each other first or remain as separate compartments with different trafficking characteristics. Our recent live-cell imaging studies are helping to clarify these issues. Using Rab proteins as specific markers to distinguish between storage vesicles and endosomes containing GLUT4, we demonstrated that the two pools remain distinct during insulin stimulation and that it is primarily internal GLUT4 storage vesicles (GSVs) marked by Rab10 that approach the plasma membrane and fuse. Our new findings add strong support to the model that GSV release from intracellular retention plays a major role in supplying GLUT4 molecules onto the PM under insulin stimulation.
Quantifying spatial organization in point-localization super-resolution images using pair correlation analysis
The distinctive distributions of proteins within subcellular compartments, both at steady state and during signaling events, play essential roles in cell function. We developed a method for delineating the complex arrangement of proteins within subcellular structures visualized by point-localization super-resolution (PL-SR) imaging. The approach, called pair-correlation photoactivated localization microscopy (PC-PALM), uses a pair-correlation algorithm to precisely identify single molecules in PL-SR imaging data sets and is used to decipher quantitative features of protein organization within subcellular compartments, including the existence of protein clusters and the size, density, and number of proteins in these clusters. We provided a step-by-step protocol for PC-PALM, illustrating its analysis capability for four plasma membrane proteins tagged with photoactivatable GFP (PAGFP). The experimental steps for PC-PALM can be carried out in three dimensions, and the analysis can be done in about 6–8 h. Researchers require substantial experience in single-molecule imaging and statistical analysis to conduct the experiments and carry out this analysis.
Visualizing cell structure and function with point-localization super-resolution imaging
Fundamental to the success of cell and developmental biology is the ability to tease apart molecular organization in cells and tissues by localizing specific proteins with respect to one another in a native cellular context. However, many key cellular structures (from mitochondrial cristae to nuclear pores) lie below the diffraction limit of visible light, precluding analysis of their organization by conventional approaches. Point-localization super-resolution microscopy techniques, such as PALM and STORM, are poised to resolve, with unprecedented clarity, the organizational principles of macromolecular complexes within cells, thus leading to deeper insights into cellular function in both health and disease. We have been using this technology to analyze receptor protein clustering and HIV budding from the plasma membrane.
Additional Funding
- IATAP (Intramural AIDS Targeted Antiviral Program)
Publications
- Fu D, Mitra K, Sengupta P, Jarnik M, Lippincott-Schwartz J, Arias I. Coordinated elevation of mitochondrial oxidative phosphorylation and autophagy help drive hepatocyte polarization. Proc Natl Acad Sci USA 2013;110:7288-7293.
- Sengupta P, Jovanovic-Talisman T, Lippincott-Schwartz J. Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nature Protocols 2013;8:345-354.
- Majoul I, Gao L, Betzig E, Onichtchouk D, Butkevich E, Kozlov Y, Bukauskas F, Bennett M, Lippincott-Schwartz J, Duden R. Fast structural responses of gap junction membrane domains to AB5 toxins. Proc Natl Acad Sci USA 2013;110(44):E4125–E4133.
- Chen Y, Lippincott-Schwartz J. Rab 10 delivers GLUT4 storage vesicles to the plasma membrane. Commun Integr Biol 2013;6(3):e2377.
- Sengupta P, Van Engelenburg S, Lippincott-Schwartz J. Visualizing cell structure and function with point-localization superresolution imaging. Dev Cell 2012;23:1092-1102.
Collaborators
- Irwin Arias, MD, Cell Biology and Metabolism Program, NICHD, Bethesda, MD
- Eric Betzig, PhD, Howard Hughes Medical Institute, Janelia Farm Research Campus, Ashburn, VA
- Michael Davidson, PhD, Florida State University, Tallahassee, FL
- Rainer Duden, PhD, Universität zu Lübeck, Lübeck, Germany
- Dong Fu, PhD, Cell Biology and Metabolism Program, NICHD, Bethesda, MD
- Harald Hess, PhD, Howard Hughes Medical Institute, Janelia Farm Research Campus, Ashburn, VA
- Tijana Jovanovic-Talisman, PhD, University of Hawaii at Manoa, Honolulu, Hawaii
- Michael Kozlov, PhD, Tel Aviv University, Tel Aviv, Israel
- Irina Majoul, PhD, Universität zu Lübeck, Lübeck, Germany
Contact
For more information, email lippincj@mail.nih.gov or visit lippincottschwartzlab.nichd.nih.gov.


