You are here: Home > Section on Environmental Gene Regulation
Small Regulatory RNAs and Small Proteins

- Gisela Storz, PhD, Head, Section on Environmental Gene Regulation
- Aixia Zhang, PhD, Staff Scientist
- Shantanu Bhatt, PhD, Postdoctoral Fellow
- Michael D. Dambach, PhD, Postdoctoral Fellow
- Yue Hao, PhD, Postdoctoral Fellow
- Taylor B. Updegrove, PhD, Postdoctoral Fellow
- Maureen K. Thomason, PhD, Graduate Student
- Hanbo Wang, BS, Graduate Student
- Xuefeng Yin, BS, Graduate Student
- Natasha Livingston, BS, Technical Intramural Research Training Award Fellow
- Jameice DeCoster, BS, Postbaccalaureate Intramural Research Training Award Fellow
Currently, we have two main interests: identification and characterization of small noncoding RNAs and identification and characterization of small proteins of less than 50 amino acids. Both small RNAs and small proteins have been overlooked because they are not detected in biochemical assays and the corresponding genes are poorly annotated and missed in genetic screens. However, mounting evidence suggests that both classes of these small molecules play important regulatory roles.
Identification and characterization of small regulatory RNAs
During the past 15 years, we have carried out several different systematic screens for small regulatory RNA genes in Escherichia coli. The screens, which have included computational screens for conservation of intergenic regions and direct detection after size selection or co-immunoprecipitation with the RNA–binding protein Hfq, are all applicable to other organisms. We are now examining small RNA expression using deep sequencing to further extend our identification of small RNAs, particularly antisense RNAs.
A major focus of the group has been to elucidate the functions of the small RNAs that we and others identified. Early on, we showed that the OxyS RNA, whose expression is induced in response to oxidative stress, acts to repress translation through limited base pairing with target mRNAs. We discovered that OxyS action depends on the Sm-like Hfq protein, which functions as a chaperone to facilitate OxyS RNA base pairing with its target mRNAs. We also found that another abundant and broadly conserved small RNA mimics the DNA structure of an open promoter and modulates RNA polymerase activity.
It is now clear that Hfq–binding small RNAs, which act through limited base pairing, are integral to many different stress responses in E. coli. We extensively characterized Hfq (1) and Hfq–binding determinants on target mRNAs (2). We also elucidated the important contributions of several Hfq–binding small RNAs to regulatory networks. For example, we found that FnrS, whose expression is induced by FNR upon a shift from aerobic to anaerobic conditions, acts to down-regulate the levels of a variety of mRNAs encoding metabolic enzymes not needed in the absence of oxygen. Another example is the Spot 42 RNA, whose levels are highest when glucose is present and which plays a broad role in catabolite repression by directly repressing genes involved in central and secondary metabolism, redox balancing, and the consumption of diverse non-preferred carbon sources. Many of the genes repressed by Spot 42 are transcriptionally activated by the global regulator CRP. Given that CRP represses Spot 42, the regulators participate in a specific regulatory circuit called a multi-output feedforward loop. We found that the loop can reduce leaky expression of target genes in the presence of glucose and can maintain repression of target genes under changing nutrient conditions.
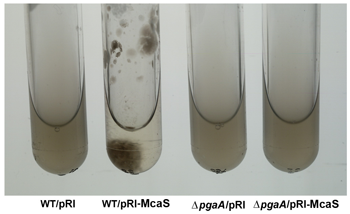
Click image to enlarge.
Figure 1. The McaS small RNA induces auto-aggregation of cells.
While cells carrying the vector control (first and third tubes from left) remain suspended, cells overexpressing the McaS small RNA (second tube from left) stick to the walls and sediment to the bottom of the test tube in the absence of shaking. The phenotype is attributable to McaS activation of the pgaABCD operon required for the synthesis and export of poly-b-1,6-N-acetyl-D-glucosamine (PGA) and thus is not is not observed in the pgaA deletion strain (fourth tube from left).
In more recent work, we discovered that the McaS RNA, whose levels are elevated in stationary phase or when glucose is limiting, regulates mRNA targets involved in various aspects of biofilm formation (3). McaS represses csgD, the transcription regulator of curli biogenesis, and activates flhD, the master transcription regulator of flagella synthesis, leading to increased motility. McaS also regulates pgaA, a porin required for the export of the polysaccharide poly β-1,6-N-acetyl-d-glucosamine. Consequently, high levels of McaS result in augmented biofilm formation while a strain lacking mcaS shows reduced biofilm formation. Based on these observations, we propose that McaS modulates steps in the progression to a sessile lifestyle.
While we assumed that sRNAs act solely by one mechanism, we discovered, to our surprise, that McaS acts by two different mechanisms: base pairing and protein titration (4). McaS base-pairs with the csgD and flhD mRNAs respectively, resulting in down-regulation and up-regulation of the corresponding cell-surface structures. In contrast, McaS activates pgaA by binding to the global RNA–binding protein CsrA, a negative regulator of pgaA translation. The McaS RNA bears at least two CsrA–binding sequences, and inactivation of these sites compromises CsrA binding, PGA regulation, and biofilm formation. Moreover, ectopic McaS expression leads to induction of two additional CsrA–repressed genes encoding diguanylate cyclases. Thus, McaS is a dual-function sRNA with roles in the two major post-transcriptional regulons controlled by the RNA–binding proteins Hfq and CsrA.
In addition to small RNAs that act via limited base pairing, we have been interested in small antisense RNAs that have the potential to form extensive base-pairing interactions with their mRNA targets encoded on the opposite strand. We previously showed that base pairing of GadY with the gadXW mRNA encoded on the opposite strand results in processing, giving rise to two halves that accumulate to higher levels than the full-length mRNA. Multiple enzymes, including the double-strand RNA–specific endoribonuclease RNase III, are involved in the GadY–directed cleavage. We also reported that a large class of antisense RNAs act to repress the synthesis of small toxic proteins. For example, in characterizing the Sib RNAs, which are encoded by five repeats in E. coli K-12, we observed an overexpression phenotype reminiscent of plasmid addiction. Further examination of the SIB repeat sequences revealed conserved open reading frames encoding highly hydrophobic 18–19 amino acid proteins (Ibs) opposite each sib gene. The Ibs proteins were found to be toxic when overexpressed, and the toxicity could be prevented by co-expression of the corresponding Sib RNA. Computational screens, together with experimental validation, showed that such small hydrophobic protein-antisense RNA gene modules, termed type 1 toxin-antitoxin modules, are much more widely distributed among bacteria than previously appreciated.
Studies to further characterize other Hfq–binding RNAs and antisense RNAs and to elucidate the roles of small RNAs that act in ways other than base pairing are ongoing.
Identification and characterization of small proteins
In our genome-wide screens for small RNAs, we found that several short RNAs do encode small proteins. The correct annotation of the smallest proteins is one of the biggest challenges of genome annotation, and perhaps more importantly, only a limited number of annotated short ORFs have been confirmed to correspond to synthesized proteins. Although such proteins have been largely missed, the few small proteins that have been studied in detail in bacterial and mammalian cells have been shown to have important functions in signaling and in cellular defenses. We thus established a project to identify and characterize E. coli proteins of less than 50 amino acids.
We used sequence conservation and ribosome binding-site models to predict genes encoding small proteins, defined as having 16–50 amino acids, in the intergenic regions of the E. coli genome. We tested expression of these predicted as well as previously annotated small proteins by integrating the sequential peptide affinity (SPA) tag directly upstream of the stop codon on the chromosome and assaying for synthesis using immunoblot assays. The approach confirmed that 20 previously annotated and 18 newly discovered proteins of 16–50 amino acids are synthesized. Studies to detect the synthesis of other predicted small proteins identified by the mapping of ribosome binding sites on a genome-wide basis (ribosome profiling) are ongoing.
Remarkably, more than half the newly discovered proteins are predicted to be single transmembrane proteins, an observation that prompted us to examine the localization, topology, and membrane insertion of the small proteins. Biochemical fractionation showed that, consistent with the predicted transmembrane helix, the small proteins generally are most abundant in the inner membrane fraction. We found examples of both Nin-Cout and Nout-Cin orientations in assays of topology-reporter fusions to representative small transmembrane proteins. Interestingly, three of nine tested proteins even display dual topology. Positive residues close to the transmembrane domains are conserved, and mutational analysis of one small protein, YohP, showed that the positive inside rule applies equally to single transmembrane domain proteins as it does, as was previously established, to larger proteins. Also, through fractionation analysis of small protein localization in strains depleted of the Sec or YidC membrane insertion pathways, we found differential requirements. Some small proteins appear to be affected by both Sec and YidC depletion, others showed more dependence on one or the other insertion pathway, while one protein was not affected by depletion of either Sec or YidC. Thus, despite their diminutive size, small proteins display considerable diversity in topology, biochemical features, and insertion pathways.
To elucidate the functions of the small proteins, we now are employing many of the approaches the group previously used to characterize the functions of small regulatory RNAs. Systematic assays for the accumulation of tagged versions of the proteins demonstrated that many small proteins accumulate under specific growth conditions or after exposure to stress. We also generated and screened bar-coded null mutants and identified small proteins required for resistance to cell envelope stress and acid shock. In addition, we are exploiting the attached sequential peptide affinity tag to identify co-purifying complexes. The combination of these approaches is giving insights into when, where, and how the small proteins act.
For example, we showed that expression of a 42–amino acid protein, now denoted MntS (formerly the small RNA gene rybA) is repressed by manganese through MntR. Overproduction of MntS causes manganese sensitivity, while a lack of MntS perturbs proper manganese-dependent repression of another manganese-regulated gene. Based on these results, we propose that MntS plays a novel role in intra-cellular manganese trafficking and homeostasis.
We also found that the 49–amino acid inner membrane protein AcrZ (formerly named YbhT), associates with the AcrAB-TolC multidrug efflux pump, which confers resistance to a wide variety of antibiotics and other compounds in E. coli (5). Co-purification of AcrZ with AcrB in the absence of both AcrA and TolC, two-hybrid assays, and suppressor mutations indicate that the interaction occurs through the inner membrane protein AcrB. The highly conserved acrZ gene is co-regulated with acrAB through induction by the MarA, Rob, and SoxS transcription regulators. In addition, mutants lacking AcrZ are sensitive to many of, but not all, the antibiotics transported by AcrAB-TolC. The differential antibiotic sensitivity suggests that AcrZ may enhance the ability of the AcrAB-TolC pump to export certain classes of substrates.
This work, together with our studies of other small proteins, suggests that many small proteins act as regulators of larger protein complexes.
Publications
- Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol;425:3678-3697.
- Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J 2012;31:1961-1974.
- Thomason MK, Fointaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 2012;84:17-35.
- Jørgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 2013;27:1132-1145.
- Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. Conserved small protein associates with the AcrB efflux pump and differentially affects antibiotic resistance. Proc Natl Acad Sci USA 2012;109:16696-16701.
Collaborators
- Susan Gottesman, PhD, Laboratory of Molecular Biology, NCI, Bethesda, MD
- Poul Valentin-Hansen, PhD, University of Southern Denmark, Odense, Denmark
Contact
For more information, email storz@helix.nih.gov or visit cbmp.nichd.nih.gov/segr.

