You are here: Home > Section on Macromolecular Recognition and Assembly
Intermolecular Forces, Recognition, and Dynamics

- Donald C. Rau, PhD, Head, Section on Macromolecular Recognition and Assembly
- Nina Yu Sidorova, PhD, Staff Scientist
- Rocco Ferrandino, BA, Postbaccalaureate Fellow
- Brandon Hoover, BA, Postbaccalaureate Fellow
- Mallory Iseminger, Special Volunteer
Our basic goal is to elucidate the coupling of the forces, structure, and dynamics of biologically important complexes and assemblies. The next challenge in structural biology is to understand the physics of interactions between molecules in aqueous solution. The ability to take advantage of the increasing number of available protein and nucleic acid structures will depend critically on establishing the link between structures and energies. A fundamental and quantitative knowledge of intermolecular forces is necessary for understanding the strength and specificity of interactions among biologically important macromolecules that control cellular function and for rationally designing agents that can effectively compete with those interactions associated with disease. We directly measure forces between biological macromolecules in macroscopic condensed arrays using osmotic stress and x-ray scattering. Our results have shown that experimentally measured forces at close spacings are different from those predicted by current, conventionally accepted theories. We interpret the observed forces as indicating a dominating contribution from water-structuring energetics.
Our practical goal is to apply the lessons from direct force measurements to the recognition and assembly reactions that control cellular processes. DNA assembly and compaction is a universal feature of cell biology. The DNA in sperm nuclei is packed densely enough that interhelical spacings can be directly measured by x-ray scattering. During vertebrate spermatogenesis, chromatin is dramatically reorganized in developing spermatids through replacement of histones with protamines to achieve compact DNA packaging. Given that DNA repair is absent in sperm, dense packaging of DNA in sperm nuclei is considered necessary to protect the DNA against damage by mutagens and reactive oxidizing species (ROS). DNA damage has been shown to correlate with infertility and likely to contribute to miscarriages and birth defects. A current objective is to establish correlations among the DNA packaging density in sperm nuclei, protamine dysfunction, and DNA damage.
The observation of the importance of hydration for intermolecular reactions also led us to investigate differences in water sequestered by complexes of sequence-specific DNA binding proteins bound to different DNA sequences, with particular emphasis on correlating binding energy and water incorporated and on the energy necessary to remove hydrating water from complexes. We determine differences in sequestered water between complexes through the effect of changing osmotic pressure on binding constants or dissociation rates. Differences in water between specific and nonspecific complexes connect thermodynamics and structure. This emphasis on water allows us to approach recognition reactions differently from standard practice. We take advantage of the unique experimental tools we developed to investigate the coupling of structure, thermodynamics, and function of these complexes from a fresh perspective.
DNA packaging in sperm nuclei
Protamines are small, arginine-rich, nuclear proteins that condense the spermatid genome into a genetically inactive state. Unlike histone-compacted DNA, protamines assemble DNA into close packed hexagonal arrays. DNA packing densities can be determined readily by x-ray scattering. The physical properties of reconstituted of protamine–DNA assemblies closely resemble those of DNA condensed by multivalent cations such as divalent manganese, cobalt hexammine, spermidine, spermine, and several arginine and lysine peptides. They are much smaller, have much less charge, and have been well characterized by us. The forces measured for compacting salmon sperm nuclei, reconstituted salmon protamine DNA arrays, and bull sperm nuclei using the osmotic stress coupled with x-ray diffraction show very similar characteristics to DNA precipitated by multivalent ions. We hypothesize that male infertility from protamine deficiency and from errors in modification is attributable to looser packing of DNA in sperm nuclei than is optimal, allowing greater accessibility to mutagens and oxidizing species.
The equilibrium surface separations of the DNA double helices compacted with the smaller multivalent ions or protamines are well defined and vary between 7 Å and 18 Å, depending on the identity of the condensing ion. The finite separation of helices indicates a delicate balancing of short-range repulsive forces with longer-range attractions. Using combined osmotic stress and single-molecule tweezing experiments, we previously characterized the distance dependence of the two component forces. The attractive force varies with DNA–DNA spacing as a 4–5 Å decay–length exponential, whereas the repulsive force is a 2–2.5 Å decay–length exponential. The difference in decay lengths by a factor of two indicates that the attractive force results from a direct interaction of charged groups on apposing helices, whereas the repulsive force is an image-charge or its hydration equivalent force. Using this constraint, we can effectively separate the attractive and repulsive contributions to the force-distance curves. Although the physical origins of the forces are still debated, we argued for water-structuring or hydration forces on the basis of our previous extensive measurements of forces between both charged and uncharged molecules.
Click image to enlarge.
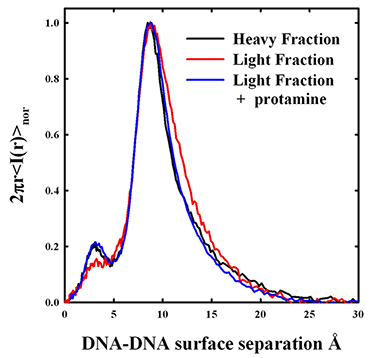
Figure 1. X-ray scattering peaks from density gradient–fractionated salmon sperm nuclei
The x-ray scattering intensity profiles are shown for sucrose density gradient–fractionated salmon sperm nuclei. The heavier fraction has a narrower distribution of interhelical spacings with a DNA-DNA surface separation of about 9 Å. The less dense fraction has the same peak spacing, but the shape of the scattering peak is different. The two fractions overlap at the smaller distances, but the least dense fraction has a DNA population subset with significantly larger spacings. The DNA with the larger spacings would be more accessible to mutagens and oxiding radicals. The addition of excess salmon protamine to the light fraction results in compaction of the more loosely packed DNA. The amplitude of the smaller scattering peak at about 3 Å is an indicator of the degree of lateral ordering in the DNA array.
We systematically investigated the contributions of protamine amino acid composition to the component forces. All vertebrate protamines use arginines almost exclusively over lysines. We found that arginine peptides can assemble DNA to much tighter spacings than lysine peptides of the same length. Packaging helices with protamines based on lysines in sperm nuclei would leave the DNA more vulnerable to damage. Not only is the attractive force component stronger with arginine than for lysine, but shorter-range repulsion is also weaker. Using synthetic arginine homo-peptides of different lengths, we found that the attractive force amplitude increases with the inverse of the number of arginines. The shorter-range repulsive force is almost independent of peptide length. Also, using synthetic peptides, we further quantitated the effect of incorporating neutral amino acids into hexa-arginine peptides. The fraction of arginines in vertebrate protamines varies from about 50–55% for mammals to about 65–70% for fish. As the fraction of arginine decreases in these synthetic peptides, the amplitude of the short-range repulsive force increases while the attraction force is only slightly affected. We hypothesize that these neutral amino acids increase the image charge–like repulsive force by displacing water from DNA grooves. With our data, the packing density of DNA in salmon sperm nuclei can be quantitatively predicted from the 21 arginines and the 33% fraction of neutral amino acids of salmon protamine.
Density fractionation of sperm is now standard practice in artificial reproduction technologies. It was found by others that the lighter fractions of density-fractionated sperm have more DNA damage than the heavier fractions. We fractionated salmon sperm nuclei on a sucrose gradient and measured x-ray interhelical scattering peaks, as shown in Figure 1. More slowly sedimenting, less dense nuclei do have a larger average spacing between helices than the heavier fractions. Careful analysis of the scattering peaks indicates that the increased average spacing is the result of an increased spread of interhelical distances from normal to larger distances rather than a simple shift of a scattering peak with constant width to larger spacings. We are thus the first to show that DNA packing densities actually vary from sperm to sperm. The addition of excess salmon protamine to the less dense nuclei results in x-ray scattering peaks that are indistinguishable from the denser nuclei. At least part of the poorer packaging of the less dense fraction is the result of insufficient protamine. We investigated the dependence of interhelical spacing on the protamine-to-DNA ratio in reconstituted assemblies. Our results indicate that the lighter-density sperm nuclei fractions may have only about 90% of the protamine necessary for normal packing densities. We are continuing to optimize a quantitative assay for protamine/DNA ratios in sperm nuclei.
We also found that salmon sperm nuclei also contain a small (1–2%) fraction of residual phosphorylated protamine. The replacement of histones by protamines occurs in several steps. Protamines are initially serine-phosphorylated as they bind to DNA. Only after removing the phosphate groups does DNA becomes tightly packed. We have evidence that serine-phosphorylation of protamines has a profound effect on DNA packing. The incorporation of one negatively charged aspartate or phosphorylated serine into a hexa-arginine peptide increases the amplitude of the short-range repulsion dramatically, the equivalent of decreasing the apparent arginine fraction to about 40–45%. The longer-range attraction is moderately reduced, but is consistent with a net +5 charge. We are now investigating whether the less dense salmon sperm nuclei fractions also have more residual protamine phosphorylation than average. We are also in the process of isolating enough phosphorylated protamine to reconstitute DNA assemblies and measure DNA spacings by x-ray.
Click image to enlarge.
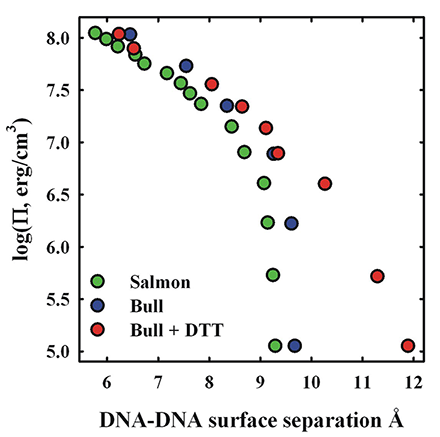
Figure 2. Comparison of salmon and bull sperm nuclei osmotic stress force curves
The osmotic pressure (Pi) of polyethylene glycol (PEG) is used to apply a force on DNA packaged in bull or salmon sperm nuclei. Consequent DNA–DNA spacings are measured by x-ray scattering. Despite the marked difference in the fraction of arginine, there is little difference in compaction forces between bull and salmon sperm nuclei. Limited reduction of bull sperm nuclei with DTT leads to a 30% increase in the spacing between DNA surfaces, consistent with the predicted effect of decreasing the fraction arginine in bull protamine relative to salmon protamine.
Many mammals have two protamines, P1 and P2; both are longer than piscine protamines. We are now investigating DNA packaging in bull sperm. Bulls only have a P1 protamine and represent an intermediate in complexity between fish and human sperm. Bovine P1 has only 26 arginines of a total of 50 amino acids. On the basis of our results with synthetic peptides, the difference in arginine content between bull (52%) and salmon (67%) protamines would be expected to result in significantly less dense compaction of DNA in mammalian sperm than in fish. A looser packaging of DNA owing to a lower arginine fraction would increase accessibility of ROS to the DNA. Bull sperm nuclei give fairly strong interhelical x-ray reflections. The peak widths, however, are broader than for salmon sperm nuclei, suggesting more disorder. In spite of the difference in arginine content, however, the osmotic stress force curve for native bull sperm nuclei is remarkably similar to that for salmon sperm nuclei (Figure 2). In both systems the equilibrium DNA–DNA spacing is 29 Å or about 9 Å of water-filled space between DNA surfaces. This can be explained by the other major difference between mammalian and piscine protamines. Extensive intra- and inter-protamine disulfide bridges are present in mammalian sperm, but absent in fish. Bull protamine P1 contains seven cysteines, whereas salmon protamine has none. Four of the cysteines in bull protamine are involved in intra-peptide disulfide bonds and the other three in inter-protamine disulfide bonds. The intra-protamine crosslinks form hairpins at the C- and N- terminals, reducing the effect length from 50 to 42 amino acids. The effective arginine fraction increases from 52% to 62%, a fraction that is now more piscine-like. Reduction of disulfide bonds with DTT appears to be a two-step process. Limited reduction of bull sperm nuclei inter-protamine disulfide bridges with 1 mM DTT leads to a 30% increase in the surface-to-surface separation of DNA helices or a 70% increase in the volume accessible to ROS (Figure 2). The increased spacing is consistent with that expected from the reduced arginine fraction of bull protamine in the absence of inter-protamine disulfide bonds. Disulfide bridges in mammals are required for tight DNA packaging, overcoming the decreased fraction arginine. The osmotic stress force curve for DTT–reduced bull nuclei gradually merges at higher pressures with the unmodified bull sperm nuclei curve (Figure 2). Significantly, disulfide bonds are not reformed if reducing DTT is removed, but will reform if both DTT is removed and nuclei are compacted with osmotic stress.
Reduction of the disulfide bonds with 10 mM DTT leads to dramatic swelling of the nuclei (at least 10–20 fold in volume) and complete loss of x-ray order. We suspect that both intra- and inter-protamine disulfide bonds are now reduced. The disulfide bonds are absolutely essential for dense packing of DNA. Once the sperm nucleus enters the egg, disulfide reduction will result in significant unpacking of the DNA. Reducing agents as glutathione have been shown necessary for decondensation of mammalian sperm nuclei after fertilization.
Future work will include: (i) coupling the structural measurements with measurements of oxidative DNA damage; (ii) density fractionation and x-ray analysis of bull sperm nuclei; and (iii) further characterization of disulfide bonds in bull sperm nuclei, in particular their re-formation after DTT reduction.
Publications
- Canchi DR, Jayasimha P, Rau DC, Makhatadze GI, Garcia AE. Molecular mechanism for the preferential exclusion of TMAO from protein surfaces. J Physical Chemistry B 2012;116:12095-12104.
- Sidorova NY, Scott T, Rau DC. DNA concentration dependent dissociation of EcoRI: direct transfer or reaction during hopping. Biophysical J 2013;104:1296-1303.
- Lander GC, Johnson JE, Rau DC, Potter CS, Carragher B, Evilevitch A. DNA bending induced phase transition of encapsidated genome in phage lambda. Nucleic Acids Res 2013;41:4518-4524.
- Derouchey JE, Hoover B, Rau DC. A comparison of DNA compaction by arginine and lysine peptides: a physical basis for arginine rich protamines. Biochemistry 2013;52:3000-3009.
Collaborators
- Jason DeRouchey, PhD, University of Kentucky, Lexington, KY
- Alex Evilevitch, PhD, Carnegie Mellon University, Pittsburgh, PA
- V. Adrian Parsegian, PhD, University of Massachusetts, Amherst, MA
- Xiangyun Qiu, PhD, George Washington University, Washington, DC
Contact
For more information, email raud@mail.nih.gov.

