You are here: Home > Section on Molecular Endocrinology
Receptors and Actions of Peptide Hormones and Regulatory Proteins in Endocrine Mechanisms

- Maria L. Dufau, MD, PhD, Head, Section on Molecular Endocrinology
- Junghoon Kang, PhD, Postdoctoral Fellow
- Mingjuan Liao, PhD, Staff Fellow
- Xiolan Sun, PhD, Postdoctoral Fellow
- Chon-Hwa Tsai-Morris, PhD, Staff Scientist
- Joaquin Villar, PhD, Postdoctoral Fellow
- Lisheng Dai, Predoctoral Fellow
We investigate the molecular basis of peptide hormone control of gonadal function, with particular emphasis on the structure and regulation of the luteinizing hormone receptor (LHR) and prolactin (PRL) receptor (PRLR) genes. We also investigate the regulatory mechanism(s) involved in the progress of spermatogenesis and the control of Leydig cell function. Our studies focus on the regulation of human LHR transcription (nuclear orphan receptors, epigenetics, DNA methylation, and second messengers, repressors, corepressors, and coactivators), as well as on the multiple-promoter control of hPRLR gene transcription. We are elucidating the functions of two inhibitory short forms of the prolactin receptors and their relevance to physiological regulation and breast cancer. We also investigate novel gonadotropin-regulated genes of relevance to the progression of testicular gametogenesis, Leydig cell function, and other endocrine processes. We specifically concentrated our studies on the function and regulation of gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25), an essential post-transcriptional regulator of spermatogenesis that was discovered, cloned, and characterized in our laboratory. The various functions of GRTH/ DDX25 provide a fertile ground for the development of a male contraceptive.
The luteinizing hormone receptor (LHR)
The luteinizing hormone receptor is a member of the G protein–coupled receptor family and is essential for sexual development and reproduction in mammals. The LHR is predominantly located on the plasma membrane of gonadal cells, where it mediates the LH signals that regulate ovarian granulosa/luteal and testicular Leydig cell development and function. It is also found in non-gonadal cells, tumoral tissues, and cancer cells, and these have provided a convenient model in which to study its transcription, which is subjected to several modes of regulation. LHR gene transcription in stable cultures of cancer cells that express the LHR is subject to repression/derepression through complex and diverse networks that include an epigenetic modulation at the promoter and association/dissociation of multiple effectors centered in SP1 at the proximal Sp1 site of the promoter. Local chromatin changes resulting from histone acetylation and cell-specific CpG island methylation/demethylation within the promoter are critical for silencing and derepression of the LHR gene in cancer cells. Chromatin changes induced by the HDAC inhibitor TSA cause cell-specific release of phosphatases that associate directly with Sp1 or indirectly through HDAC1/2 at the promoter. This serves as an "on" switch for Sp1 phosphorylation induced by PI3-K/PKCzeta at Ser 641 that triggers release of p107 repressor from SP1 at the promoter and marked transcriptional activation of the LHR gene. Maximal derepression of the LHR gene upon TSA treatment is dependent on complete demethylation of the promoter in conjunction with histone hyperacetylation and release of the repressors p107 and HDAC/mSin3A from SP1. After we had defined the role of repressor/corepressors and their association/dissociation with SP1 in TSA-induced repression/derepression of the LHR, we initiated studies on the involvement of transcriptional activators in this process. After exclusion of a number of coactivators in the Sp1-directed basal and TSA-induced transcriptional activation, including p300, CBP and PCAF and others, we investigated the participation of PC4, a potential coactivator of Sp1, in the transcriptional control of LHR. Knockdown of PC4 by siRNA inhibited basal and TSA-induced LHR gene transcriptional activation and expression in MCF7 cells. While over-expression of PC4 alone had no effect on the LHR gene, it significantly enhanced SP1- but not SP3-mediated LHR transcriptional activity. PC4 directly interacts with SP1 at the LHR promoter; this interaction is negatively regulated by PC4 phosphorylation. The coactivator domain of PC4 and DNA binding domain of Sp1 are essential for PC4/SP1 interaction. ChIP assays revealed significant occupancy of PC4 at the LHR promoter, which increases upon TSA treatment. Disruption of PC4 expression significantly reduced TSA-induced recruitment of TFIIB and RNAPII to the promoter. Moreover, our findings demonstrated that the critical role of PC4 in TSA-induced LHR gene expression is independent of SP1 phosphorylation and release of corepressors, indicating its role as linker coactivator of Sp1 and one or more components of the transcriptional machinery. These recent studies have demonstrated a crucial aspect of LHR gene modulation whereby PC4 acts as an essential coactivator of Sp1 to facilitate LHR transcription.
Gonadotropin-regulated testicular RNA helicase (GRTH/DDX25)
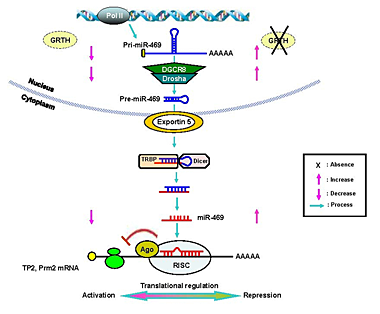
Figure 1. GRTH/DDX25 is required for the structural and functional integrity of the chromatoid body
Panel I. Electron microscopic analysis of the chromatoid body (CB) structure present in round spermatids isolated from wild-type mice (WT) control (typical structure) (white arrow) or abnormal structure after incubation of cells with the inhibitor of nuclear export leptomycin B (LMB) or of mRNA transcription (5,6-dichloro -1-β-D-ribofuranosyl-benzimidazole-DRB) for 3 or 6 h (striped arrows). N, nucleus. Panel II. CB from spermatids of WT and GRTH/Ddx25 knockout (KO) testis. Panel III. The diagram illustrates the relevance of GRTH for the structural integrity of the CB (hexagon) in spermatids of the mouse testis. Distribution of GRTH/ Ddx25 (black diamond), Mouse Vasa Homolog (MVH/Ddx4) (triangle), and MIWI (striped circle) in cellular compartments of round spermatids. Abnormally compact CB noted in round spermatids after treatment with inhibitors LMB or DRB resembles the CB in GRTH KO mice. MVH and MIWI, members of RNA Induced Silencing Complex, are excluded from the CB in the KO and in round spermatids cultured in presence of LMB or DRB. (click image to enlarge)
GRTH is a multifunctional enzyme that was discovered in our laboratory. It is an essential regulator of sperm maturation and an excellent target for the development of a male contraceptive. GRTH prevents cell apoptosis. A missense mutation of the GRTH/DDX25 gene, associated with absence of phosphorylated GRTH cytoplasmic species, was found in 5.8% of infertile patients with azoospermia. GRTH null mice are sterile due to spermatid arrest and failure to elongate. Through its association with CRM-1, GRTH participates in the export of selected messages relevant to the progress of spermatogenesis (chromatin remodeling proteins: transition protein 1 and 2, protamine 1 and 2; cytoskeleton structural proteins: FSc1 and Odf1; and testicular angiotensin enzyme and others) from nuclear to cytoplasmic sites, including the chromatoid body (CB) of spermatids (equivalent to somatic P bodies, contains members of the RNA-induced silencing complex [RISC] and is viewed as a storage/processing site of mRNAs during spermatogenesis) and polyribosomes, where it participates in translation of proteins that are essential for spermatogenesis. The presence of GRTH in the CB of the wild-type (WT) mice and the major structural changes and size changes observed in the round spermatids of the GRTH null mice indicated that GRTH's functions take place, at least in part, in the CB. Our recent studies provided direct evidence for a nuclear/cytoplasmic shuttling function of GRTH in germ cells. We demonstrated the cellular localization of GRTH in spermatids during various stages of the spermatogenesis of WT mice at nuclear, cytoplasmic sites and specifically in the CB. Further, we determined changes in relevant cytoplasmic and CB proteins observed in the GRTH-null mice which, unlike GRTH, are not found at nuclear sites or participate in transport in the GRTH WT mice. Such proteins include the mouse Vasa homolog (MVH), a RNA helicase distantly related to GRTH, which concentrates in the CB and interacts with Dicer in vitro, and MIWI, a murine PIWI/Argonaut protein that has been proposed to be a member of RISC, with residence in the CB. Both proteins are excluded from the abnormal CB of GRTH null mice and accumulate only in the cytoplasm. To address the relevance of transport function of GRTH and associated messages to the structure of CB, we used acute cultures of round spermatids of WT mice. Treatment of these cells with either an inhibitor of nuclear export or one of RNA synthesis caused nuclear retention of GRTH and its absence from the cytoplasm and CB. On other hand, proteins of the RISC complex were excluded from the CB and accumulated in the cytoplasm upon treatment with either inhibitor, as occurred in spermatids of KO mice. EM studies demonstrated that the CB changed from a large lobular-filamentous structure to a small condensed one after treatment, resembling the CB of the GRTH-KO. GRTH did not interact with RISC members. Thus, rather than participating in translational regulation, the nuclear export of GRTH-RNP complexes might be required to maintain the CB structural integrity in spermatids and to deliver essential RNAs to the CB for silencing/storage and/or degradation of genes during germ cell development. In other studies we demonstrated the involvement of GRTH in the export of its own message from the nucleus to the cytoplasm. Association of GRTH message with GRTH protein was observed in testis extracts of WT mice. Significant nuclear accumulation of GRTH message, reflected by increases in the ratio of nuclear to cytoplasmic RNA level, were observed upon blockade of export with inhibitor in cultured germ cells. These studies demonstrated the essential participation of the GRTH export/transport function of the CB and for the integrity of functional components within this organelle during spermatogenesis (Figure 1).
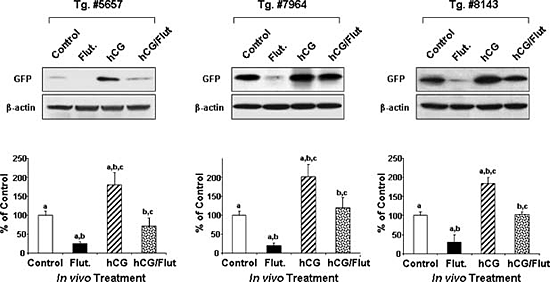
Figure 2. Gonadotropin/androgen regulation of GRTH transcription in transgenic mice
Gonadotropin/androgen-mediated action in 1kb (−1085/+63 bp) 5'-flanking region of GRTH-GFP transgenic mice. Top panel. Western blot analysis using anti-GFP antibody to assess GFP expression in the testis of three transgenic mice lines after in vivo hormone (500 ng hCG) treatment for 24 hr. Also, animals were pretreated with an androgen receptor antagonist (Flutamide -Flut, 5mg twice a day/3days) prior to hCG treatment to inhibit basal and hCG-stimulated androgen action. Lower panel. Summary of analysis normalized by β-actin. Results are presented as percent of individual treatment relative to control sample. hCG treatment caused a significant increase in GFP protein expression compared with untreated control. Flutamide markedly decreased the basal (endogenous androgen) and abolished hCG/androgen-stimulated GFP expression. The studies demonstrated that the 1 kb 5'-flanking region of the GRTH gene contains sequences for androgen regulation of the gene's expression in Leydig cells.
GRTH is the only member of the RNA helicase family of proteins that is known to be hormonally regulated. GRTH in Leydig cells (LC) is transcriptionally regulated by hCG in vitro and in vivo via cAMP by androgen and androgen receptors (AR) present in Leydig cells (LC). We envisage a paracrine regulation of GRTH expression in germ cells resulting from the androgen/AR in Sertoli cells. To define basal and hormonally regulated transcription of the GRTH gene and the regulatory regions required for cell-specific gene expression, we generated transgenic mice carrying deletions of the GRTH 5'-flanking sequence upstream of the promoter domain, with GFP as the reporter gene (GFP-reporter). Our findings indicated that cell-specific expression of the GRTH gene is differentially directed by its 5'-flanking sequence. GFP-reporter gene expression directed by the GRTH 5'-flanking sequences extending to −3.6 kb was specifically located in Leydig cells, and the 205 bp minimal promoter domain was sufficient for this cell-specific expression. The absence of expression in the tubular compartment observed in this study indicated that the GRTH germ cell–targeted sequence(s) (pachytene spermatocytes, spermatids) is/are localized upstream to 3.6 kb of the GRTH 5'-flanking region (from 3.6 kb to 6.4 kb). The −6.4 kb region of GRTH is followed upstream by the coding sequences of the uridine synthetase gene, which are unlikely to direct GRTH expression to germ cells. We are currently generating transgenic mice containing a −6.4kb/+63bp construct to elucidate germ cell–specific targeting sequences in the GRTH gene. There may be specific cis requirement(s) to direct the GRTH gene in different testicular cells. Upstream interactions with specific transfactors at downstream sites could be required for GRTH expression in germ cells. The regulatory mechanism for cell-specific differential utilization of 5'-flanking sequences in the GRTH gene will be studied next. In vivo, treatment by hCG markedly increased GFP expression directed by the 1 kb (5' to the ATG codon) transgene. Administration of the androgen receptor (AR) inhibitor Flutamide blocked basal and hCG-stimulated GFP expression in LC. This indicated that the 1 kb fragment of the GRTH gene contains sequences for androgen regulation of its expression in LC (Figure 2). In follow-up studies in vitro and in vivo, we localized an ARE half-site at −800, which binds to AR, as the functional site for gonadotropin-induced androgen action on GRTH transcription/expression.
Prolactin receptor (PRLR)
The prolactin receptor (PRLR) is a member of the lactogen/cytokine receptor family, which mediates the diverse cellular actions of PRLR in several target tissues. The PRL is plays major role in the proliferation and differentiation of breast epithelium and is the primary hormone involved in the stimulation and maintenance of lactation. It is also a tumor promoter in rodents and has been implicated in the development of breast cancer. Our laboratory demonstrated that human hPRLR expression is controlled by a complex regulatory system at the transcriptional level that is governed by multiple promoters. These include the preferentially utilized generic promoter 1/exon1 (hPIII/hEI3), which is also present in rat and mouse and five human-specific exon-1/promoters (hE1N1-5). E2 regulates PRLR expression through the PIII promoter via a non-classical ERE-independent mechanism in target cells. The E2/ERα complex translocates to the nucleus and interacts with C/EBPβ and SP1 bound to cognate elements on the hPIII, leading to recruitment of coactivators to the complex, with consequent region-specific changes in histone acetylation. These hormone receptor–induced associations and chromatin changes favor TFIIB and RNA polymerase II recruitment, leading to activation of hPIII-directed transcription and PRLR gene expression.
In recent studies, we mapped the physical domains involved in interactions between ERα and transcription factors SP1 and C/EBPβ. Direct protein interaction of ERα with SP1 and C/EBPβ is essential for up-regulation of PRLR transcription by E2. The basic region and leucine zipper of C/EBPβ, zinc fingers of SP1 and DNA binding domain of ERα, were identified by pull-down assays as the responsible regions for the interactions between ERα, C/EBPβ, and SP1 in vitro. Simultaneous or sequential addition of SP1 and/or C/EBPβ provided evidence regarding the assembly of these transfactors. Compared with SP1, C/EBPβ interacted weakly with ERα within the complex (shown in simultaneous incubation). The efficiency of the recruitment of C/EBPβ was highly enhanced when ERα/Sp1 complexes were preformed in sequential studies. This led us to propose that the association of ERα and SP1 enhances the recruitment of C/EBPβ into the complex. In addition to C/EBPβ association with the complex through ERα, our studies have indicated a strong direct interaction between C/EBPβ and Sp1, which may serve to stabilize the ERα/Sp1/C/EBPβ complex. BRET assays and complementation analysis established that the ERα dimer and E2/ERα complexes participate in the interaction with transcription factor Sp1 and C/EBPβ. Specifically, BRET assays in live cells demonstrated interactions of ERα/ERα, ERα/C/EBPβ, and ERα/SP1, which were significantly increased by estradiol. In addition, interactions between C/EBPβ/SP1, SP1/SP1, and C/EBPβ/CEBPβ were observed. BRET, with a bimolecular complementation assay applied to ERα, revealed that ERα exists as a constitutive homodimer and that E2 binding induced change(s) in the ERα homodimer conformation favorable for its association with C/EBPβ and SP1 mono- and/or homodimers. Our studies indicate that the enhanced complex formation of the ERα dimer with SP1 and C/EBPβ plays an essential role in the transcriptional activation of the hPRLR gene.
Publications
- Xie YL, Hassan SA, Qazi AM, Tsai-Morris CH, Dufau ML. Intramolecular disulfide bonds of the prolactin receptor short form are required for its inhibitory action on the function of the long form of the receptor. Mol Cell Biol. 2009;29:2546-2555.
- Sheng Y, Tsai Morris C-H, Li J, Dufau ML. Lessons from the gonadotropin-regulated long chain acyl-CoA synthetase (GRLACS) null mouse model: a role in steroidogenesis, but not result in X-ALD phenotype. J Steroid Biochem Mol Biol. 2009;114:44-56.
- Dufau ML, Liao M, Zhang Y. Participation of signaling pathways in the derepression of luteinizing hormone receptor transcription. Mol Cell Endocrinol. 2010;314:221-227.
- Sato H, Tsai-Morris CH, Dufau ML. Relevance of gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) in the structural integrity of the chromatoid body. Biochim Biophys Acta. 2010;1803:534-543.
- Tsai-Morris CH, Sheng Y, Gutti R, Li J, Pickel J, Dufau ML. Gonadotropin-Regulated testicular RNA helicase (GRTH/DDX25) gene: cell-specific expression and transcriptional regulation by androgen in transgenic mouse testis. J Cell Biochem. 2010;109:1142-1147.
Collaborators
- Sergio A. Hassan, PhD, Center for Molecular Modeling, CIT, NIH, Bethesda, MD
- James Pickel, PhD, Laboratory of Genetics, Transgene Core, NIMH, Bethesda, MD
Contact
For more information, email dufau@helix.nih.gov.


