You are here: Home > Section on Neuroendocrinology
Pineal Gland, Chronobiology, and Neuroepigenetics

- David C. Klein, PhD, Head, Section on Neuroendocrinology
- Diego Bustos, PhD, Postdoctoral Fellow
- Sam Clokie, PhD, Postdoctoral Fellow
- Steven L. Coon, PhD, Staff Scientist
- M.A.A. Namboodiri, PhD, Guest Researcher
- Margaret Ochocinska, PhD, Postdoctoral Fellow
- Jiri Pavlicek, PhD, Postdoctoral Fellow
- Harvey Pollard, MD, PhD, Guest Researcher
- Martin Rath, Special Volunteer
- Louise Rovsing, Special Volunteer
- Joan L. Weller, BA, Senior Research Assistant
The Section on Neuroendocrinology studies the pineal gland and regulation of the pineal hormone melatonin. The work has broad implications for vertebrate biology and is of special interest to clinical scientists studying human diseases relating to circadian rhythms, including endocrine pathologies, sleep and mood disorders, and deficiencies in alertness. The Section also addresses the factors that control the remarkable global changes in gene expression occurring in the pineal gland on a 24-hour basis as well as factors associated with the establishment of the pineal phenotype. The scope of interest covers the range of small and large non-coding RNAs, in addition to mRNAs. The Section is dedicated to advancing understand of pineal function through the use of high-throughput sequencing to examine the pineal transcriptome.
The timezyme: arylalkylamine N-acetyltransferase
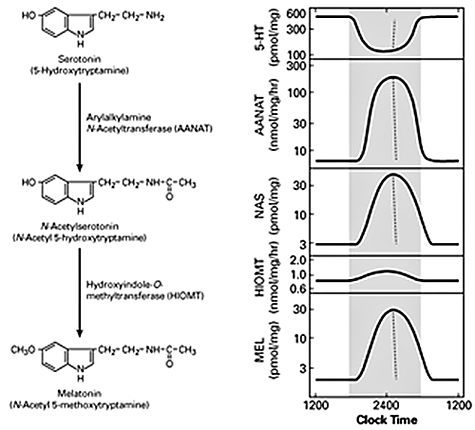
Figure 1. Daily rhythm in indole metabolism in the pineal gland
The daily rhythm in circulating melatonin production reflects the increased production in the pineal gland, as depicted here. During the day, melatonin production is low due to the low levels of AANAT. At night, AANAT activity rises, resulting in an increase in N-acetylserotonin. The increase in N-acetylserotonin raises melatonin production by a mass action effect; the level of the last enzyme in melatonin synthesis is constant. Changes in melatonin synthesis cause parallel changes in melatonin release. Circulating melatonin is rapidly destroyed in the liver, which allows changes in production and release to be immediately reflected in changes in circulating melatonin.
A pivotal advance in our investigations into arylalkylamine N-acetyltransferase (AANAT), the second enzyme in the pathway for melatonin synthesis from serotonin, was our finding that the enzyme is critical to the control of the rhythm in melatonin synthesis (Figure 1). In all species examined to date, the large increase in melatonin synthesis at night causes an increase in the production of melatonin, which is a highly conserved feature of vertebrate physiology.
The transcriptional mechanisms that control expression of the AANAT gene include interactions of cyclic AMP–response elements in the promoter region; other response elements in the promoter, including the E-box and photoreceptor conserved elements, appear to determine the marked tissue-specific pattern of expression of the gene, which is limited to the pineal gland and retina. We determined that, in rodents, expression of AANAT rises about 100-fold at night; however, this feature of regulation is not highly conserved. We discovered little or no nocturnal increase in AANAT expression in ungulates and primates. Thus, transcriptional control mechanisms are not essential for regulating melatonin synthesis in all vertebrates.
With regard to highly conserved post-translational control of AANAT, the workers in the Section and their collaborators determined that it is mediated by cyclic AMP and involves phosphorylation at C- and N-terminal sites by cyclic AMP–dependent protein kinase. This leads to formation of a reversible complex with 14-3-3 protein, in which the enzyme is stabilized and activated, as indicated by structural, in vivo and in vitro studies. When cyclic AMP levels fall, AANAT is dephosphorylated and destroyed by the proteasome.
Molecular evolution of AANAT
We focused on an important element of the AANAT molecule—a floppy loop described as Loop 1, which forms a portion of the arylamine-binding pocket. Loop 1 has undergone notable changes during the course of the enzyme's evolution. In collaboration with Daniel Sackett and Sergio Hassan, we evaluated the importance of specific residues in the floppy loop to determine their contribution to the greater than 1,000-fold increase in specific activity of AANAT that has occurred during chordate evolution, based on studies of Amphioxus that were carried out by Pascaline Gaildrat and Jack Falcón. During the course of chordate evolution, Loop 1 acquired the tripeptide CPL65, which is responsible for increased enzyme activity; removal of the CPL tripeptide markedly reduced activity. Moreover, increasing the local flexibility of the tripeptide region by P64G and P64A mutations had a counterintuitive effect, reducing both activity and the overall movement of Loop 1, as estimated from Langevin dynamics simulations. Binding studies indicate that the mutations increased the off-rate constant of a model substrate without altering the dissociation constant. The structural kink and local rigidity imposed by Pro-64 may enhance activity by favoring configurations of Loop 1 that facilitate catalysis without becoming immobilized by intramolecular interactions.
Studies conducted in collaboration with Jack Falcón revealed that there was a rapid expansion of the invertebrate type of AANAT in Amphioxus, resulting in seven copies. In addition, it was found that only the invertebrate form of the enzyme is present in this group. Studies on the eel and dogfish reveal the opposite—these chordates have only the vertebrate form of the enzyme. However, a striking exception was the discovery that the elephant fish, a member of the chimera sharks, has both vertebrate and invertebrate forms.
Neuroepigenetics: global control of daily changes in pineal gene expression are regulated by an adrenergic/cyclic AMP mechanism.
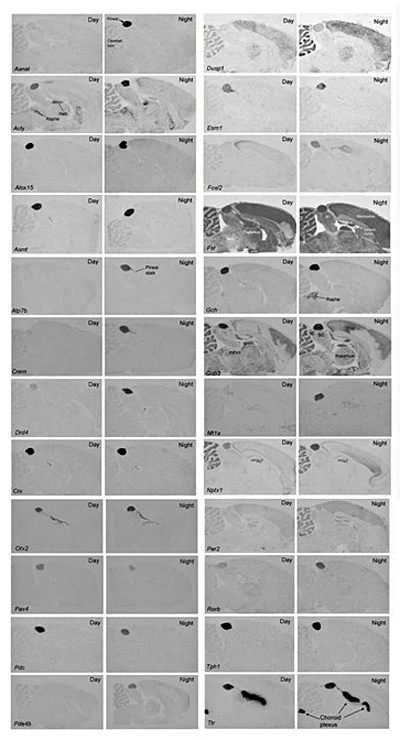
Figure 2. Gene expression in the rat pineal gland: night/day differences and tissue specificity
Each panel contains images of in situ hybridization autoradiographs prepared from sagittal sections of rat brains through the pineal gland. The sections on the left are from animals killed during the day and those on the right are from animals killed during the night. The sections were incubated with antisense probes identified in the bottom left-hand corner of the Day image. Hab, habenula; ic, inferior colliculus; mhn, medial habenular nucleus; Raphe, dorsal raphe nucleus; sc, superior colliculus. These figures are available in high resolution at sne.nichd.nih.gov. For further details see the original report (Bailey et al., 2009).
We initiated several projects aimed at 1) obtaining a global picture of differences in gene expression that occur on a night/day basis and 2) identifying genes that are highly enriched in the pineal gland. Together with Igor Dawid, we facilitated the NICHD-sponsored commercialization of an Affymetrix® chip for analysis of gene expression in zebrafish and Xenopus. In collaboration with Dawid, Yoav Gothilf, and Reiko Toyama, we are characterizing pineal gene expression in the zebrafish and other species as a function of time of day and of development. We already identified a set of genes highly expressed in the pineal gland. In collaboration with Peter Munson, David Carter, and Ruben Baler, we are analyzing the control of gene expression in second-messenger cascades. The work has triggered several investigations that analyze the expression of genes that previously had not appeared in the pineal literature, including those encoding methionine adenosyl transferase, MAP kinase phosphatase-1, PEPT1, phoshodiesterase 4D2, dopamine Drd4 receptor, and a subunit of the IgE receptor.
The group of pineal-specific genes expressed in the pineal gland of several vertebrates includes genes known to be associated with melatonin production and visual signal transduction. In addition, we identified many genes (over 600), new to the pineal literature, that exhibit more than two-fold night/day differences in expression and another set of genes that are more highly expressed in the pineal gland than in other tissues. This work is leading to a rapid increase in knowledge of the biochemical profile, conserved across species, of the pineal gland. It is also pointing to new transcriptional pathways controlled by previously unrecognized transcription factors; by analyzing these transcription factors and the promoters of genes that are either upregulated at night or highly expressed in the pineal gland, we will be able to construct a regulatory network that describes the cascade of transcription factors underlying the control of pineal gene expression. We also identified potentially new and important pathways involved in cell-cell communication and signal transduction in the pineal gland; expression of some genes points to new functions for the pineal gland.
Using quantitative RT-PCR and radiochemical in situ hybridization (Figure 2), our collaborators David Sugden, Morten Møller, and Martin Rath confirmed the results of microarray studies on the rat pineal gland. This approach has provided a more detailed and quantitative description of the 24-hour patterns of gene expression, revealing that the patterns are not all similar or in phase but rather exhibit novel features. Our results permit a full-scale biochemical and physiological analysis of the control of genes in the pineal gland. Results from organ culture work revealed that the vast majority of genes that exhibit night/day changes in expression are also regulated by norepinephrine, acting through a cyclic AMP mechanism that may involve cyclic AMP–response element binding sites in regulated genes and less specific epigenetic mechanisms (Figure 3). The daily changes in gene expression in the pineal gland impact many processes, as outlined in Figure 3.
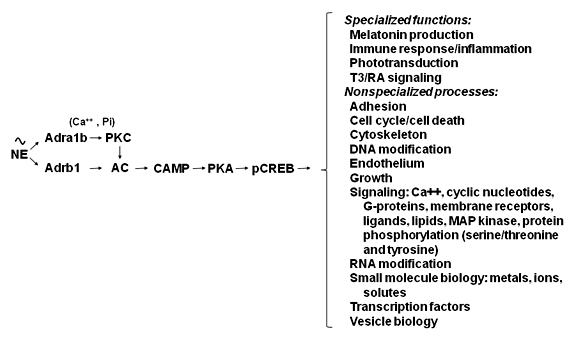
Figure 3. Neural regulation of gene expression in the rat pineal gland
Sympathetic nerves release norepinephrine (NE), which binds to β1-adrenergic receptors (Adrb1)and to α1b-adrenergic receptors (Adra1b). NE acts through the Adrb1 receptor to partially activate adenylate cyclase (AC) and through Adra1b to potentiate activation of the Adrb1 receptor. This interaction involves Adra1b-dependent activation of Ca++ and phosphatidyl inositol (Pi) activation of protein kinase C (PKC), which enhances Adrb1 stimulation of AC. The resulting increase in cyclic AMP(CAMP) leads to activation of protein kinase A(PKA), which phosphorylates cyclic AMP response element binding protein(CREB). The consequence of this is the global shift in gene expression as illustrated in Figures 1 and 2. The transcription factor profile unique to this tissue establishes which genes are regulated by CREB phosphorylation.
This line of investigation has now been extended using high-throughput/next-generation sequencing, which has confirmed many, but not all the indications from microarray. In addition, this effort identified over one hundred genes that are expressed in a 24-hour pattern in the pineal gland, revealed previously unrealized expressed introns and flanking regions, and identified a set of highly tissue-specific and rhythmically expressed non-coding RNAs. The findings will form the foundation of future studies on mechanisms involved in neural control of gene expression.
Transcriptional control by NeuroD-1, CRX, OTX, Pax4, Pax6, and Mbnl2
Our microarray analyses, which were confirmed with independent techniques, revealed that the pineal gland expresses high levels of several transcription factors, including NeuroD1, CRX OTX, Pax4, and Pax6, that play a role in the development of the eye and other tissues. With collaborators under the direction of Morten Møller, we undertook a detailed description of the developmental appearance of these transcripts; our long-term plan is to determine the transcripts' role in developmental and adult expression of genes in the pineal gland.
The vertebrate homeobox gene Otx2 is essential for the development of rostral brain regions. It also appears to play a role in the development of retinal photoreceptor cells and pinealocytes. We demonstrated the temporal expression pattern of Otx2 in the rat brain, with exceptionally strong expression in the pineal gland throughout late embryonic and postnatal stages. Widespread high expression of Otx2 in the embryonic brain becomes progressively restricted in the adult to the pineal gland. Crx (cone-rod homeobox), a downstream target gene of Otx2, showed a pineal expression pattern similar to that of Otx2, despite a distinct lag in time of onset. We identified the Otx2 protein in pineal extracts and found it to be localized in pinealocytes. Total pineal Otx2 mRNA neither showed night/day variation nor was influenced by the removal of sympathetic input, suggesting that the level of Otx2 mRNA is independent of the photoneural input to the gland. Our observations are consistent with the view that pineal expression of Otx2 is required for development and plays a role in the adult by controlling the expression of the cluster of genes associated with phototransduction and melatonin synthesis.
NeuroD1/BETA2, a member of the bHLH transcription factor family, influences the fate of specific neuronal, endocrine, and retinal cells. In collaboration with Estela Muñoz, we found that NeuroD1 mRNA is highly abundant in the developing and adult rat pineal gland. Pineal expression begins in the 17-day embryo, at which time it is also detectable in other brain regions. Expression in the pineal gland increases during the embryonic period and is maintained thereafter at levels equivalent to those found in the cerebellum and retina. In contrast, NeuroD1 mRNA decreases markedly in non-cerebellar brain regions during development. Pineal NeuroD1 levels are similar during night and day and do not appear to be influenced by sympathetic neural input. Gene expression analysis of the pineal glands from neonatal NeuroD1 knockout mice identified 127 transcripts that are more than two-fold downregulated and 16 that are more than two-fold upregulated. Quantitative RT-PCR showed that the most dramatically downregulated gene is KIF5C (kinesin-family member 5C) (about 100-fold); the most dramatically upregulated gene is that encoding glutamic acid decarboxylase 1 (about four-fold). Other affected transcripts encode proteins involved in differentiation, development, signal transduction, and trafficking. To elucidate the role of NeuroD1 in the rodent pinealocyte, we are using a Cre-Lox conditional knockout strategy to block expression of NeuroD1 in both the pineal gland and retina.
Our studies revealed that Pax4 is strongly expressed in the rat pineal gland and retina. The levels of pineal Pax4 transcripts are low in the fetus and increase postnatally; Pax6, on the other hand, exhibits an inverse pattern of expression and is more strongly expressed in the fetus. In the adult, the abundance of Pax4 mRNA exhibits a diurnal rhythm in the pineal gland, with maximal levels occurring late during the light period. Sympathetic denervation of the pineal gland by superior cervical ganglionectomy prevents the nocturnal decrease in pineal Pax4 mRNA. At night, the release of norepinephrine by sympathetic innervation adrenergically stimulates the pineal gland. We found that treatment with adrenergic agonists suppresses pineal Pax4 expression both in vivo and in vitro, suggesting that pineal Pax4 function is mediated by cyclic AMP, a second messenger of norepinephrine in the pineal gland. These findings suggest that the nocturnal decrease in pineal Pax4 mRNA is controlled by the sympathetic neural pathway, which, in turn, controls pineal function via an adrenergic cyclic AMP mechanism. The daily changes in Pax4 expression may influence gene expression in the pineal gland.
Most recently, we reported that the pineal gland expresses very high levels of the gene encoding muscleblind-like 2 (Mbnl2), a zinc finger protein first identified in Drosophila. The gene appears to be essential for photoreceptor development and is involved in RNA splicing. Mbnl2 is strongly expressed in the rat pineal gland. The abundance of pineal Mbnl2 transcripts follows a marked circadian rhythm with peak levels approximately seven-fold higher at night than during the day. Mbnl2 protein exhibits a similar rhythm. In vitro studies indicate that the abundance of Mbnl2 transcripts and protein are controlled by an adrenergic/cAMP mechanism.
Regulation of dopamine Drd4R receptor: cAMP + T3 "AND" gate regulation
Preliminary cDNA microarray studies suggested that Drd4R expression increases at night. Our collaborator Morten Møller confirmed such expression by in situ analysis, as we did by Northern blot and quantitative PCR. Other than trace levels, we could not find expression of other dopamine receptors in the pineal gland. Drd4R expression in the pineal gland is many-fold higher than in most other tissues, suggesting that the Drd4R receptor plays an important role in pineal signal transduction, perhaps as an inhibitor of adrenergic stimulation of adenylate cyclase. Regulation of the expression of this gene is unusual; in the case of other genes studied to date, including AANAT, MAT2, MKP-1, Dio2 (which encodes type II deiodinase), ICER, and FRA-2, the nocturnal increase in mRNA can be reproduced in organ culture by treating with either norepinephrine or a cyclic AMP agonist. However, expression of Drd4R receptor mRNA does not increase following treatment with either norepinephrine or a cyclic AMP agonist. Rather, co-treatment with the thyroid hormone triiodothyronin (T3) and either norepinephrine or cyclic AMP is required to increase Drd4R mRNA, thereby establishing that expression of the Drd4R receptor is regulated by an "AND" gate operated by cAMP and T3 and providing the first evidence of a role for the thyroid gland in regulating a specific gene in the pineal gland. This finding is consistent with the knowledge of an increase in the capacity to convert thyroxin (T4) to T3 through induction of Dio2 at night in the pineal gland, indicating that melatonin synthesis might be a function of thyroid status. Our study thus has broad implications for dopamine signal transduction in other tissues.
As part of a broader effort to understand the similarities between the retina and pineal gland, a semiquantitative in situ hybridization study revealed prominent night/day variation in Drd4 expression in the retina of the rat, with a peak during the night. Drd4 expression is seen in all retinal layers, but the nocturnal increase is confined to the photoreceptors. Removal of sympathetic input to the eye does not affect retinal Drd4 expression, but T3 treatment induces Drd4 expression in the photoreceptors. In a developmental series, we found that expression of Drd4 is restricted to postnatal stages, with a peak at postnatal day 12. The high Drd4 expression in rat retinal photoreceptors during the night supports physiological and pharmacological evidence that the Drd4 receptor is involved in the dopaminergic inhibition of melatonin synthesis upon light stimulation. The sharp increase of Drd4 expression at a specific postnatal time suggests that dopamine is also involved in retinal development.
Additional Funding
- University of Copenhagen
- United States - Israel Binational Science Foundation grant numbers 2009290 and 2005280
Publications
- Kim JS, Coon SL, Weller JL, Blackshaw S, Rath MF, Møller M, Klein DC. Muscleblind-like 2: circadian expression in the mammalian pineal gland is controlled by an adrenergic-cAMP mechanism. J Neurochem. 2009;110:756-764.
- Bailey MJ, Coon SL, Carter DA, Humphries A, Kim JS, Shi Q, Gaildrat P, Morin F, Ganguly S, Hogenesch JB, Weller JL, Rath MF, Møller M, Baler R, Sugden D, Rangel ZG, Munson PJ, Klein DC. Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. J Biol Chem. 2009;284:7606-7622.
- Pavlicek J, Sauzet S, Besseau L, Coon SL, Weller JL, Boeuf G, Gaildrat P, Omelchenko MV, Koonin EV, Falcón J, Klein DC. Evolution of AANAT: expansion of the gene family in the cephalochordate amphioxus. BMC Evol Biol. 2010;10:154.
- Klein DC, Bailey MJ, Carter DA, Kim JS, Shi Q, Ho AK, Chik CL, Gaildrat P, Morin F, Ganguly S, Rath MF, Møller M, Sugden D, Rangel ZG, Munson PJ, Weller JL, Coon SL. Klein DC, Bailey MJ, Carter DA, Kim JS, Shi Q, Ho AK, Chik CL, Gaildrat P, Morin F, Ganguly S, Rath MF, Møller M, Sugden D, Rangel ZG, Munson PJ, Weller JL, Coon SL. Pineal function: impact of microarray analysis. Pineal function: impact of microarray analysis. Mol Cell Endocrinol. 2010;314:170-183.
- Kim JS, Bailey MJ, Weller JL, Sugden D, Rath MF, Møller M, Klein DC. Thyroid hormone and adrenergic signaling interact to control pineal expression of the dopamine receptor D4 gene (Drd4). Mol Cell Endocrinol. 2010;314:128-35.
Collaborators
- Michael J. Bailey, PhD, Texas A&M University, College Station, TX
- Ruben Baler, PhD, Laboratory of Cellular and Molecular Regulation, NIMH, Bethesda, MD
- Seth Blackshaw, PhD, The Johns Hopkins University, Baltimore, MD
- David Carter, PhD, University of Wales, Cardiff, UK
- Praveen Cherukuri, PhD, Genome Technology Branch, NHGRI, Rockville, MD
- Constance L. Chik, MD, University of Alberta, Edmonton, Canada
- Igor Dawid, PhD, Program on Genomics of Differentiation, NICHD, Bethesda, MD
- Jack Falcón, PhD, Observeratoire Oceanologique/CNRS, Banyuls-sur-Mer, France
- Takaishi Furukawa, PhD, Osaka Bioscience Institute, Osaka, Japan
- Pascaline Gaildrat, PhD, Faculté de Médecin, Centre Hospitalier Universitaire, Rouen, France
- Sandra Goebbels, PhD, Max-Planck-Institut für Experimentelle Medizin, Göttingen, Germany
- Yoav Gothilf, PhD, Tel Aviv University, Tel Aviv, Israel
- Sergio A. Hassan, PhD, Center for Molecular Modeling, CIT, NIH, Bethesda, MD
- Anthony Ho, PhD, University of Alberta, Edmonton, Canada
- John Hogenesch, PhD, Genome Institute of Novartis Foundation, San Diego, CA
- P. Michael Iuvone, PhD, Emory University School of Medicine, Atlanta, GA
- Howard Jaffe, PhD, Laboratory of Neurochemistry, NINDS, Bethesda, MD
- Jong-So Kim, PhD, Pohang University of Science and Technology, Pohang, South Korea
- Ken Kirk, PhD, Laboratory of Chemistry, NIDDK, Bethesda, MD
- Eugene V. Koonin, PhD, National Center for Biotechnology Information, NLM, Bethesda, MD
- Morten Møller, PhD, Panum Institutet, Københavns Universitet, Copenhagen, Denmark
- Fabrice Morin, PhD, Université de Rouen, Mont-Saint-Aignan, France
- Jim Mullikin, PhD, Genome Technology Branch, NHGRI, Rockville, MD
- Estela Muñoz, PhD, Instituto de Histología y Embriología, Universidad Nacional de Cuyo, Mendoza, Argentina
- Peter Munson, PhD, Mathematical and Statistical Computing Laboratory, CIT, NIH, Bethesda, MD
- Tomas Obsil, PhD, Charles University, Prague, Czech Republic
- Zoila Rangel, MS, Mathematical and Statistical Computing Laboratory, CIT, NIH, Bethesda, MD
- Martin F. Rath, PhD, Panum Institutet, Københavns Universitet, Copenhagen, Denmark
- Daniel Sackett, PhD, Program on Physical Biology, NICHD, Bethesda, MD
- Jürgen Schnermann, MD, Kidney Disease Branch, NIDDK, Bethesda, MD
- David Sugden, PhD, Kings College, University of London, London, UK
- Larry Szewczuk, PhD, The Johns Hopkins University, Baltimore, Maryland
- Reiko Toyama, PhD, Program on Genomics of Differentiation, NICHD, Bethesda, MD
- David Weaver, PhD, University of Massachusetts, Worcester, MA
- Pin-Xian Xu, PhD, Mount Sinai School of Medicine, New York, NY
- Al Yergey, PhD, Biomedical Mass Spectrometry Facility, NICHD, Bethesda, MD
Contact
For more information, email kleind@mail.nih.gov or visit sne.nichd.nih.gov.


