You are here: Home > Section on Neurophysiology and Biophysics
Structural Biology of the Glutamate Receptor

- Mark L. Mayer, PhD, Head, Section on Neurophysiology and Biophysics
- Sagar Chittori, PhD, Visiting Fellow
- Janesh Kumar, PhD, Visiting Fellow
- Suvendu Lomash, PhD, Visiting Fellow
- Anthony Berger, BSc, Postbaccalaureate Fellow
- Carla Glasser, PhD, Technical Specialist
Ionotropic glutamate receptors (iGluRs) are membrane proteins that act as molecular pores permeable to sodium and calcium ions. At the majority of excitatory synapses in the mammalian nervous system, iGluRs mediate rapid signal transmission, converting the pre-synaptic action potential–triggered release of glutamate into a depolarizing post-synaptic potential. The seven iGluR gene families in humans encode 18 subunits, which assemble to form three major functional families named after the ligands that were first used to identify iGluR subtypes in the late 1970s: AMPA, kainate, and NMDA. Given the essential role of iGluRs in normal brain function and development, and mounting evidence that dysfunction of iGluR activity mediates several neurological and psychiatric diseases and damage during stroke, we devote substantial effort to analyzing iGluR function at the molecular level. Atomic resolution structures solved by protein crystallization and X-ray diffraction provide a framework for designing electro-physiological and biochemical experiments aimed at defining the mechanisms underlying ligand recognition, the gating of ion channel activity, and the action of allosteric modulators. Information derived from these experiments will permit the development of subtype-selective antagonists and allosteric modulators with novel therapeutic applications and reveal the inner workings of a complicated protein machine that plays a critical role in brain function.
Key issues in the field include obtaining structures for iGluRs trapped in their three major conformational states, i.e., the resting, activated, and desensitized states, and obtaining insight into the energy landscapes connecting these states. Also of interest are the evolutionary relationships connecting iGluRs in different species and how structurally related chemosensors bind to a wide range of small molecules.
Expression, purification, and structural analysis of full-length kainate receptor ion channels
Structural studies on CNS membrane proteins are notoriously difficult because commonly used bacterial protein expression systems cannot be employed to express eukaryotic membrane proteins. We recently succeeded in obtaining highly purified preparations of two different kainate receptor subtypes expressed in both insect cells, using baculovirus expression, and HEK 293 cells, using transient transfection. For both systems, cultures must be grown on the 12–liter and larger scale in order to obtain sufficient protein for biochemical and structural analysis. Because the amounts of protein obtained are still at least an order of magnitude lower than for soluble proteins, it is necessary to use highly efficient methods for establishing conditions in which the proteins remain, for several days following purification, correctly folded and do not aggregate or dissociate into dimers and monomers. To do this, we use tryptophan fluorescence size exclusion chromatography (FSEC), for which only microgram quantities of purified protein are required for each run of a 300 mM–length analytical gel filtration column. Using this approach, we screened numerous detergents, lipids, receptor ligands, and mutants with selective removal of glycosylation sites and various C-terminal deletions. Crystallization trails using conventional detergent-solubilized protein, as well as bicelles and lipidic cubic phases, are under way, with initial hits of microcrystals that are candidates for optimization. Proteins that we prepared using this approach were also used successfully to solve 25 Å resolution structures for GluK2 in resting and desensitized states.
Structural studies on the amino-terminal domain of iGluRs
Glutamate receptor ion channels (iGluRs) are excitatory neurotransmitter receptors with a unique molecular architecture, in which the extracellular domains assemble as a dimer of dimers. The structure of individual dimer assemblies was established previously for both the isolated ligand-binding domain (LBD) and more recently for the larger amino-terminal domain (ATD). How these dimers pack to form tetrameric assemblies in intact iGluRs has remained controversial. Adding to the complexity, native glutamate receptor ion channels are tetrameric assemblies containing two or more distinct subunits. While some AMPA and kainate receptors can form functional homomeric ion channels, the KA1 and KA2 subunits are obligate heteromers, which function only in combination with GluR5-7; in the brain the major kainate receptor species contains both GluR6 and KA2 subunits in unknown stoichiometry and geometric arrangement. The mechanisms controlling glutamate receptor assembly are believed to involve an initial step in which the ATDs assemble as dimers, but how the GluR6 and KA2 subunits cooperate to do this had not been established. Using sedimentation velocity analysis, we found that the ATDs of GluR6 and KA2 coassemble as a heterodimer of Kd 11 nM, which is 32,000-fold lower than the Kd for homodimer formation by KA2. We then solved crystal structures for the GluR6/KA2 ATD heterodimer and heterotetramer assemblies. Using these structures as a guide, we performed a mutant cycle analysis to probe the energetics of assembly and showed that high-affinity ATD interactions are required for biosynthesis of functional heteromeric receptors. The high affinity of the KA2 subunit for GluR6 ensures that ATD heterodimers form early during the process of biogenesis, before trafficking comes into play, and in addition provides a mechanism by which formation of functional GluR6 homotetramers that lack the KA2 subunit is suppressed, while ensuring a 2:2 stoichiometry of assembly. The binding mechanism generating the kainate receptor heterodimer assembly involves residues present in both the R1 and R2 lobes of KA2 protomers.
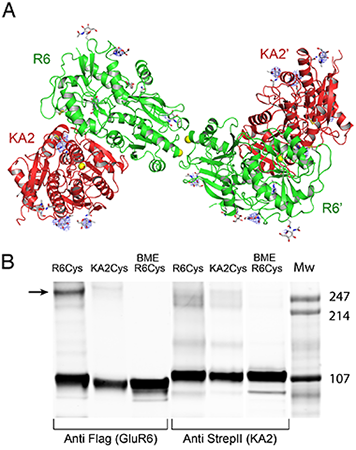
Figure 1. Crystal structure of the GluR6 KA2 ATD heterotetramer assembly
(A) Crystal structure of the GluR6/KA2 heterotetramer ATD assembly for wild-type GluR6 and KA2, colored green and red respectively; the ribbon diagram shows one of three identical tetramers for the 10 protomers in the asymmetric unit, with one tetramer formed by non-crystallographic symmetry operations; side chains that support N-linked glycosylation are drawn as sticks; yellow spheres indicate the positions in the GluR6 and KA2 subunits at which cysteine mutations were introduced to test for formation of disulfide cross links in full length receptors. Electron density maps (Fo-Fc contoured at 3 s blue mesh) illustrate features corresponding to glycan residues, which were not included in the model or used for refinement. (B) Western blots run under non-reducing conditions for detergent-solubilized, affinity-purified (StrepII tag) full-length heteromeric GluR6/KA2, in which Cys mutants were introduced at equivalent sites in the domain R2 lateral surface of either GluR6 or KA2 and probed against Flag (GluR6) and StrepII (KA2) epitopes; lanes 3 and 6 contain the same samples loaded in 1 and 4 but with the addition of 10 mM beta-mercaptoethanol (BME).
Molecular biophysical studies on AMPA receptor dimer assembly
Click image to enlarge.
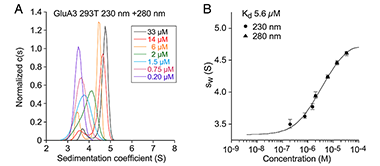
Figure 2. Sedimentation analysis of the AMPA receptor GluA3 subunit ATD
Sedimentation velocity AUC analysis for the GluA3 ATD. (A) Normalized sedimentation coefficient c(s) distributions measured at 230 and 280 nm at the protein concentrations indicated, from 0.2 to 33 µM, plotted in units of experimental s-values. (B) The sw isotherm derived by integration fit with model for monomer-dimer association.
Sedimentation velocity (SV) analytical ultracentrifugation (AUC) has re-emerged as an important tool in the characterization of biological macromolecules and has been used extensively for analysis of monomer-dimer and dimer-tetramer equilibria for the ATDs and LBDs of iGluRs. Prior reports on AMPA receptor GluA2 dimerization differed in their estimate of the monomer-dimer Kd by a 2,400-fold range, with no consensus as to whether the ATD forms tetramers in solution. We found by sedimentation velocity (SV) analysis, performed using absorbance detection, a narrow range of monomer-dimer Kd values for GluA2, from 5–11 nM for six independent experiments, with no detectable formation of tetramers, and no effect of glycosylation or the polypeptide linker connecting the ATDs and LBDs; for GluA3 the monomer-dimer Kd was 5.6 µM, again with no detectable tetramer formation. For sedimentation equilibrium (SE) experiments we obtained a wide range of Kd values for GluA2, from 13–284 nM, while for GluA3 (Kd of 3.1 µM) there was a less than two-fold difference from the SV value. After the one-week centrifuge run, analysis of cell contents by silver stained gels revealed low MW GluA2 breakdown products. Simulated data for SE runs demonstrate that the apparent Kd for GluA2 varies with the extent of proteolysis, leading to artificially high Kd values. SV experiments with fluorescence detection for GluA2 labeled with 5,6-carboxyfluorescein, and fluorescence anisotropy measurements for GluA2 labeled with DyLight405, yielded Kd values of 5 and 11 nM, consistent with those from SV with absorbance detection. However, the sedimentation coefficients measured by AUC using absorbance were strikingly different from those obtained using fluorescence systems and, for the latter, are not consistent with hydrodynamic protein models. Thus, for unknown reasons the concentration dependence of sedimentation coefficients obtained with fluorescence detection SV may be unreliable, limiting the usefulness of this technique for quantitative analysis. Given the unique capabilities of the fluorescence detection AUC in future experiments, we plan to investigate this in detail, with the goal of identifying procedures that permit accurate Kd measurements at nM protein concentrations.
Structural studies on glutamate receptors from primitive organisms
We expressed the LBD of a recently discovered glutamate receptor from the primitive eukaryote Adineta vaga (AvGluR1) in Origami B (DE3) and purified it to homogeneity. The apo form of AvaR1 was generated for a direct filter-binding assay, yielding a Kd of 200 nM. Competition assays reveal that AvGluR1 has promiscuous ligand-binding activity, and in addition to recognizing acidic and neutral amino acids, binds also to alanine, methionine, and phenylalanine, but not to amino acids with a beta carbon branch, such as valine and threonine. Electrophysiological assays establish that, like glutamate and aspartate, alanine, methionine, and phenylalanine are efficient agonists. Crystal structures solved to investigate this unusual binding pattern reveal that the gamma COOH group of glutamate and aspartate is coordinated by by a pair of arginine residues in domain 2, unlike in any other iGluR LBD structure solved to date, but similar to the mechanisms for binding of glutamate by type III GPCRs. Neutral and hydrophobic amino acids use a chloride ion as a surrogate ligand, compensating for the missing ligand carboxylate group. Consistent with this, competition assays with tritiated glutamate reveal that the binding of alanine and serine, but not of glutamate and aspartate to AvGluR1, is chloride dependent.
Structural basis for allosteric modulation of kainate receptors by zinc
Kainate receptors (KARs) play a key role in the regulation of synaptic networks. We find that zinc, a cation released at a subset of glutamatergic synapses, potentiates glutamate currents mediated by homomeric and heteromeric KARs containing GluK3 at 10–100 µM concentrations, whereas it inhibits other KAR subtypes. Potentiation of GluK3 currents is mainly due to reduced desensitization, as shown by kinetic analysis and modeling. Mutation and crystallographic analyses revealed that a specific zinc-binding site is formed at the base of the LBD dimer interface by a GluK3–specific aspartate (Asp759), together with two conserved residues, His762 and Asp730, the latter located on the partner subunit. This site of interaction identifies the LBD as a hot spot for allosteric modulation, given that kainate receptors are also modulated by sodium and chloride ions, which bind to discrete sites at the top of the dimer assembly, while AMPA–receptor allosteric modulators bind in a crevice between subunits in their LBD dimer assembly. Our analysis suggests that tetrameric GluK2/3 receptors are likely assembled as pairs of heterodimeric LBDs. Therefore, zinc binding stabilizes the labile GluK3 dimer interface, slows desensitization, and potentiates currents, providing a new mechanism for KAR potentiation at glutamatergic synapses.
Additional Funding
- NICHD IRP
Publications
- Mayer ML. Emerging models of glutamate receptor ion channel structure and function. Structure 2012;19:1370-1380.
- Zhao H, Berger AJ, Brown PH, Kumar J, Balbo A, May CA, Casillas E Jr, Laue TM, Patterson GH, Mayer ML, Schuck P. Analysis of high-affinity assembly for AMPA receptor amino-terminal domains. J Gen Physiol 2012;139:371-388.
- Kumar J, Schuck P, Mayer ML. Structure and assembly mechanism for heteromeric kainate receptors. Neuron 2011;71:319-331.
- Das U, Kumar J, Mayer ML, Plested AJ. Domain organization and function in GluK2 subtype kainate receptors. Proc Natl Acad Sci USA 2010;107:8463-8468.
- Mayer ML. Structure and mechanism of glutamate receptor ion channel assembly, activation and modulation. Curr Opin Neurobiol 2011;21:283-290.
Collaborators
- Albert Lau, PhD, The Johns Hopkins University School of Medicine, Baltimore, MD
- Christophe Mulle, PhD, Interdisciplinary Institute for Neuroscience, Université de Bordeaux, Bordeaux, France
- Peter Schuck, PhD, Laboratory of Cellular Imaging and Macromolecular Biophysics, NIBIB, Bethesda, MD
- Sriram Subramaniam, PhD, Laboratory of Cell Biology, Center for Cancer Research, NCI, Bethesda, MD