You are here: Home > Section on Drosophila Gene Regulation
Regulation of Homeotic Gene Function in Drosophila

- James A. Kennison, PhD, Head, Section on Drosophila Gene Regulation
- Alexander Conant, Summer Intern
- Monica T. Cooper, BA, Senior Research Technician
- Mark Mortin, PhD, Staff Scientist
Our goal is to understand how genes control cell fates during development. The homeotic genes in Drosophila specify cell identities at both the embryonic and adult stages. They encode homeodomain-containing transcription factors that control cell fates by regulating the transcription of downstream target genes. The homeotic genes are expressed in precise spatial patterns that are crucial for the proper determination of cell fate. Both loss of expression and ectopic expression in the wrong tissues lead to changes in cell fate. These changes provide powerful assays for identifying the trans-acting factors that regulate the homeotic genes and the cis-acting sequences through which they act. Both the homeotic genes and the trans-acting factors that regulate them are conserved between Drosophila and human. In addition to many conserved developmental genes, at least half of the disease- and cancer-causing genes in man are conserved in Drosophila, making Drosophila a particularly important model system for the study of human development and disease.
Cis-acting sequences for transcriptional regulation of the Sex combs reduced homeotic gene
Assays in transgenes in Drosophila previously identified cis-acting transcriptional regulatory elements from the homeotic genes. The assays identified tissue-specific enhancer elements as well as cis-regulatory elements that are required for the maintenance of activation or repression throughout development. While these transgenic assays have been important in defining the structure of the cis-regulatory elements and identifying trans-acting factors that bind to them, their functions within the context of the endogenous genes remain poorly understood. We used a large number of existing chromosomal aberrations in the Sex combs reduced homeotic gene to investigate the functions of the cis-acting elements within the endogenous gene. These chromosomal aberrations identified an imaginal leg enhancer about 35 kb upstream of the Sex combs reduced promoter. This enhancer is not only able to activate transcription of the Sex combs reduced promoter that is 35 kb distant, but it can also activate transcription of the Sex combs reduced promoter on the homologous chromosome. Although the imaginal leg enhancer can activate transcription in all three pairs of legs, it is normally silenced in the second and third pairs of legs. The silencing requires the Polycomb group proteins. We are currently trying to identify the cis-regulatory DNA sequences in the Sex combs reduced gene that are required for transcriptional activation in the first leg and for Polycomb group silencing in the second and third legs. Characterization of the chromosomal rearrangements in Figure 1 also revealed that two genetic elements (proximal and distal MES) about 70 kb apart in the Sex combs reduced gene must be in cis to maintain proper repression. When not physically linked to each other, these elements interact with elements on the homologous chromosome and cause derepression of its wild-type Sex combs reduced gene. We tested DNA fragments from the Sex combs reduced gene in transgenic assays to identify endogenous cis-regulatory elements that could interact. From the regions that include the regulatory elements required for the maintenance of silencing, we identified two clusters of DNA fragments that promote pairing-sensitive silencing (an assay for interaction of cis-regulatory DNA fragments). We are using targeted gene replacement to delete these clusters of elements in the endogenous gene to assay their role in transcriptional regulation.
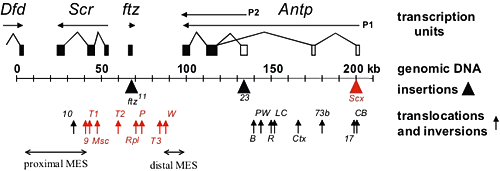
Figure 1. Chromosomal aberrations in the distal half of the Antennapedia complex
The transcription units are above the genomic DNA, while chromosomal aberrations are below (solid triangles indicate insertions of transposable elements and upward arrows indicate breakpoints of translocations and inversions). Chromosomal aberrations in red interfere with silencing in the adult second and third legs. The regions that include the proximal and distal MES are indicated by the horizontal arrows.
Trans-acting activators and repressors of homeotic genes
The initial domains of homeotic gene repression are set by the segmentation proteins, which also divide the embryo into segments. Genetic studies have identified the trithorax group of genes that are required for expression or function of the homeotic genes, including the maintenance of transcriptional activation. Maintenance of transcriptional repression requires the proteins encoded by the Polycomb group genes. To identify new trithorax group activators and Polycomb group repressors, we are screening for new mutations that mimic the phenotypes: loss of function or ectopic expression of the homeotic genes. We have generated over 4,000 lethal mutants and, among those that die late in development, have identified two dozen mutants with homeotic phenotypes. Some of the homeotic phenotypes are shown in Figure 2. These mutants identify genes required for expression or function of the homeotic genes.

Figure 2. Homeotic phenotypes of new pharate-adult lethal mutants
Panel A shows wild-type on the left and the transformation of aristae to distal leg on the right. Panel B shows a wild-type haltere on the left and transformations of anterior and posterior haltere to anterior and posterior wing in the middle and right, respectively. Panel C shows the first legs from a wild-type male on the left, and three different mutants with reduced sex combs on the right. Panel D shows a mutant male with sex combs on both the first and second tarsal segments. Panel E shows a mutant male with sex combs on all three pairs of legs. Panel F shows on the left the abdominal segments from a wild-type male and and on the right a mutant male with transformation of the fifth abdominal segment to a more anterior identity.
Reduced function of the trithorax group genes mimics loss of function of the homeotic genes. We previously identified several trithorax group genes that encode subunits of chromatin-remodeling complexes. The brahma, moira, and osa genes encode subunits of the Brahma chromatin-remodeling complex, which is conserved from yeast to human. To understand further the function of the Brahma complex, we have been isolating and characterizing mutations that interact with brahma. One of the interacting genes, female-sterile homeotic, encodes a bromodomain protein that interacts with regulatory elements near the Ultrabithorax homeotic gene promoter. The interaction appears to be important for rendering the homeotic promoter accessible to interactions with proteins bound to distant regulatory elements. Another gene that we have identified by its mutant interactions with brahma is rhinoceros. rhinoceros encodes the Drosophila homolog of a mammalian transcription factor that interacts with the von Hippel Lindau tumor suppressor. The characterization of two other brahma-interacting genes, verthandi and Pearl, suggests that two protein complexes known to be important for chromosome segregation during mitosis also play important roles in transcriptional regulation. We showed that verthandi encodes the RAD21 subunit of the cohesin complex. The cohesin complex is required for proper segregation of sister chromatids during anaphase. We also showed that Pearl encodes one of the two gamma-tubulin isoforms in Drosophila. During the characterization of Pearl, we identified a recessive suppressor mutation that rescues the zygotic lethality associated with loss of gamma-tubulin function. This recessive suppressor mutation is in a gene that encodes a protein required for formation of the gamma-tubulin ring complex. Our results demonstrate that gamma-tubulin and microtubule organizer complexes are required not only for chromosome segregation during mitosis but may also play a role in transcriptional regulation.
Publications
- Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, Hunter SM, Keall R, Warren WD, Brock HW, Sinclair DAR, Honda BM. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc Natl Acad Sci USA 2008 105:12405-12410.
- Vázquez M, Cooper MT, Zurita M, Kennison JA. γTub23C interacts genetically with Brahma chromatin-remodeling complexes in Drosophila melanogaster. Genetics 2008 180:835-843.
Collaborators
- Hugh W. Brock, PhD, University of British Columbia, Vancouver, Canada
- Dale Dorsett, PhD, Saint Louis University School of Medicine, St. Louis, MO
- Barry M. Honda, PhD, Simon Fraser University, Burnaby, Canada
- Der-Hwa Huang, PhD, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan, ROC
- Martha Vazquez, PhD, Instituto de Biotecnología, UNAM, Cuernavaca, Mexico
- Mario Zurita, PhD, Instituto de Biotecnología, UNAM, Cuernavaca, Mexico
Contact
For more information, email kennisoj@mail.nih.gov.



