You are here: Home > Unit on Behavioral Neurogenetics
Neuronal Circuits Controlling Behavior: Genetic Analysis in Zebrafish

- Harold Burgess, PhD, Head, Unit on Behavioral Neurogenetics
- Kandice Fero, PhD, Postdoctoral Fellow
- Markus C. Hannan, Postbaccalaureate Fellow
- Raul Rojas, PhD, Research Technician
- Alex Wendling, Summer Student
- Tohei Yokogawa, PhD, Postdoctoral Fellow
Our goal is to understand how neuronal circuits in larval zebrafish produce appropriate motor responses under diverse environmental contexts. Locomotor behavior in zebrafish larvae is controlled by neuronal circuits that are established through genetic interactions during development. We aim to identify genes and neurons that are required for the construction and function of the brainstem circuits underlying specific behaviors. Neuronal circuits situated in the brainstem form the core of the locomotor control network in vertebrates, responsible for balance, posture, motor control, and arousal. Accordingly, many neurological disorders stem from abnormal formation or function of brainstem circuits. Insights into the function of brainstem circuits in health and disease have come from genetic manipulation of neurons in zebrafish larvae, in combination with computational analysis of behavior.
Several unique features of the zebrafish model system facilitate analysis of the neuronal basis of vertebrate behavior. The larval zebrafish brain exhibits the basic architecture of the vertebrate brain, but is much less complex than the mammalian brain. Because of the optical clarity of the embryo, it is easy to visualize individual neurons and manipulate them using genetic techniques. Behavior in larvae is innate and therefore exhibits minimal variability between fish. Subtle alterations in behavior can therefore be robustly scored, making it possible to quickly assess the contribution of identified neurons to a variety of motor behaviors. We use two major behavioral paradigms to investigate these questions: modulation of the acoustic startle response by prepulse inhibition, and modulation of the locomotor repertoire during a phototaxis-based navigational task. In addition, we are developing a suite of genetic tools and behavioral assays to probe the nexus between neuronal function and behavior at single-cell resolution.
Modulation of the acoustic startle response
We previously demonstrated that intense acoustic stimuli elicit two types of startle response in zebrafish larvae: rapid short-latency responses and lower-performance long-latency responses. All fish can generate both types of response, but which response emerges is unpredictable from trial to trial. We aim to understand how larvae choose to deploy a short- or long-latency response. In earlier work, we found that the short-latency responses are modulated in a similar fashion to startle responses in mammals. in which startle magnitude is inhibited when the startle stimulus is preceded by a weak auditory prepulse. This form of startle modulation, termed prepulse inhibition, is diminished in several neurological conditions, including schizophrenia. Previously, we conducted a screen to identify fish carrying genetic mutations resulting in a reduction in prepulse inhibition. Using these fish, we are now performing linkage analysis to map the genetic mutations in the mutants and identify genes required for prepulse inhibition. These experiments will identify circuit components that selectively suppress generation of short-latency responses. In parallel, we are analyzing how long-latency responses are modulated. Brainstem neurons that trigger a motor response must belong to the restricted cohort of neurons that project to the spinal cord. We are therefore sequentially ablating neurons of this class using a pulsed nitrogen laser, then probing the stimulus threshold and magnitude of the long-latency startle response system. Together, these approaches will allow us to find neuronal mechanisms for the implementation of behavioral choice during startle responses in zebrafish larvae.
Functional mapping of serotonergic neuronal architecture
The serotonergic system is a major neurotransmitter system modulating behavior in higher vertebrates. To ascertain the role of this system in behavioral control in larval zebrafish, we generated transgenic fish expressing the GAL4 transcription factor in serotonergic neurons. This enables us to genetically manipulate these neurons by crossing the fish to lines carrying reporter genes under the control of the UAS promoter, for example to a UAS:Nitroreductase line to genetically ablate neurons. Consistent with our hypothesis, nitroreductase ablation of serotonergic neurons modulates a discrete subset of behaviors. We are now using two-photon based laser ablation to destroy single serotonergic neurons in vivo to identify the subset of neurons that modulate startle responses. As a second approach toward identifying cells that modulate startle, we generated transgenic fish in which the UAS promoter drives expression of mEOS-FP, a monomeric photoconvertible fluorescent protein. This system will allow us to trace the connections of single serotonergic neurons in vivo, and, in combination with our ablation studies, establish a neuronal-level functional map of serotonergic anatomy.
Tools for analyzing neuronal circuits that control behavior
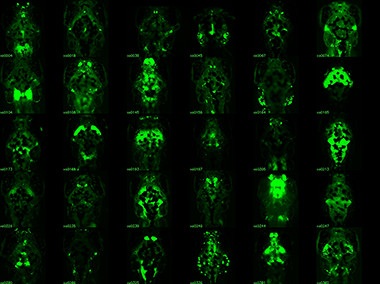
Enhancer trap lines for neurobehavioral research
Examples of transgenic Gal4 enhancer trap lines recovered screening to date. Gal4 expression is visualized by crossing lines to a fluorescent UAS:Kaede reporter. All images are dorsal views of the brain, taking advantage of the transparency of larva. Lines with broad expression in the nervous system are recovered as well as lines labeling specific structures and lines with very restricted patterns of expression. Scale bar 100 microns. (click image to enlarge)
The relatively simple nervous system of zebrafish larvae and restricted range of motor behaviors makes it possible to identify neuronal pathways that underlie the entire behavioral repertoire. For this to be feasible, it would be extremely useful to have reporter lines that would permit the manipulation of small groups of neurons known to be involved in a particular motor behavior. We are therefore performing a screen to identify neurons required for normal behavioral performance. This is a two-part process, first using an enhancer trap approach with a Gal4 reporter construct to create zebrafish lines with restricted patterns of neuronal expression. We then use these lines to genetically ablate trapped neurons and screen larvae for defects in locomotor behavior. To date, we have established more than 100 transgenic lines with unique patterns of expression of the Gal4 reporter and built an automated screening platform that allows us to rapidly screen the entire repertoire of locomotor behaviors. Preliminary data from behavioral screening with several lines revealed defects in specific behaviors. This screen will thus generate a set of reporter lines that identify and provide experimental access to cohorts of neurons linked to specific behaviors. The lines will constitute a unique resource for decoding the developmental genetics and anatomical basis of behavior in zebrafish larvae. This is the first time such a screen has been attempted in a vertebrate organism.
Publications
- Fero K, Yokogawa T, Burgess HA. The behavioral repertoire of larval zebrafish. Zebrafish Models in Neurobehavioral Research 2010;in press.
- Burgess HA, Schoch H, Granato M. Distinct retinal pathways drive spatial orientation behaviors in zebrafish navigation. Current Biology 2010;20:381-6.
- Pei W, Kratz LE, Bernardini I, Sood R, Yokogawa T, Dorward H, Ciccone C, Kelley RI, Anikster Y, Burgess HA, Huizing M, Feldman B. A model of Costeff Syndrome reveals metabolic and protective functions of mitochondrial OPA3. Development 2010;137:2587-96.
Collaborators
- Benjamin Feldman, PhD, Medical Genetics Branch, NHGRI, Bethesda, MD
- Fumihito Ono, PhD, Laboratory of Molecular Physiology, NIAAA, Rockville, MD
- David W. Raible, PhD, University of Washington, Seattle, WA
- David Valle, MD, The Johns Hopkins University School of Medicine, Baltimore, MD
- Michael Wolfgang, PhD, The Johns Hopkins University School of Medicine, Baltimore, MD
Contact
For more information, email haroldburgess@mail.nih.gov or visit ubn.nichd.nih.gov.


