You are here: Home > Section on Molecular Regulation
Global Regulation of Gene Expression by ppGpp

- Michael Cashel, MD, PhD, Head, Section on Molecular Regulation
- Tamara James, PhD, Intramural Research Training Award Fellow
Two naturally occurring nucleotide regulators, abbreviated as ppGpp and pppGpp, continue to hold our interest. These are simple analogs of GDP and GTP, whose ribose 3′ hydroxyl bears a pyrophosphate. The (p)ppGpp nucleotides are sometimes called alarmones because they signal nutritional and environmental stress in bacteria, archea, plants, and arguably now in animal cells and serve to trigger adaptive responses. They were also called magic spots because they responded like "magic" to a bewildering variety of subtle changes in central metabolism that signal stress. Synthesis of (p)ppGpp is activated when the aminoacylation of any tRNA species falls short of the demands of protein synthesis. The mechanism provides a continuous survey of the metabolic availability of each amino acid addition to nascent proteins, whether availability is governed by de novo synthesis, uptake, or catabolism. Hydrolysis of (p)ppGpp is inhibited when low steady-state levels of the acylated carrier protein (ACP) donor limit synthesis of lipids. Changes of the ratio of acyl/non-acyl ACP trigger a conformational change in the (p)ppGpp hydrolase. This circuit provides a broad sensor for subtle steady state deficiencies in carbohydrate, energy, and membrane metabolism.
Regulation by (p)ppGpp is global. It affects about one third of total gene expression, primarily by affecting transcription and secondarily at the biochemical level by indirect regulatory effects of interactions with specific enzymes. The action of (p)ppGpp on transcription is unique for bacteria because it interacts with the RNA polymerase protein itself, without participation of other sequence-recognition elements. Regulation requires an RNA polymerase–binding protein (DksA), which acts in concert to modify the kinetic properties of promoter initiation in either a positive or negative way, depending on the promoter sequence. Patterns of E. coli global regulation have emerged. During starvation, (p)ppGpp inhibits promoters for genes active during luxuriant growth (rRNA, tRNA, r-proteins), which become superfluous during starvation. Activation occurs of transcription of genes encoding stress-adaptive functions by (p)ppGpp.
The research is important because this simple signal governs the fundamental ability of cells to adjust gene expression to physiological stress. A clinical importance for (p)ppGpp has emerged because, in many instances, bacterial pathogens survive the stress of host defenses via (p)ppGpp. It is now appreciated that (p)ppGpp plays key roles in enhancing virulence and in fostering antibiotic-resistant “persistence” of many bacterial pathogens, including M. tuberculosis. Regulation by (p)ppGpp is currently being exploited to develop new antibiotics, such as "relacin," and also provides methods for dissecting stages of pathogenesis defined by their (p)ppGpp dependence. It is noteworthy that (p)Gpp–deficient attenuated bacterial vaccines have proven effective in mouse models.
How ppGpp, pppGpp works: structural and regulatory interactions with RNA polymerase.
This year, we achieved a more rigorous definition of the specificity of interactions between the RNA polymerase protein (1). The achievement required solving five challenges. First, conditions to gratuitously induce differential cellular accumulation of either ppGpp or pppGpp need to be devised such that contamination by the sister nucleotide is minimized, which allows definition of physiological effects unique to each analog. If activities are shared rather than unique, induction of one or the other allows the specific regulatory activities of each to be estimated in vivo. Second, the induction condition must occur during balanced growth and cannot itself have physiological consequences (i.e., constitute a stress) other than induction. The third challenge is to find conditions for the simple enzymatic synthesis of pure preparations of ppGpp as well as pppGpp, which allows their use as reagents to assess in vitro regulatory activities of each nucleotide. Fourth, to rule out indirect effects, transcription regulation requires conditions that allow documentation of regulation in reactions with pure components for transcription. These include pure RNA polymerase holoenzyme and DksA protein together with promoter templates able to display positive or negative (p)ppGpp regulation. The last challenge was met through a collaboration with Katsuhiko Murakami, who provided RNA polymerase holoenzyme crystals that permitted documentation of a ppGpp or pppGpp binding site.
We found induction of cellular ppGpp to be about a 10-fold more potent inhibitor of balanced growth than when pppGpp was induced. We independently substantiated this effect on growth by measuring the ratios of RNA/DNA in cells induced by ppGpp or pppGpp. An important control with inactivating missense mutants of the synthetase revealed that observed regulatory effects were attributable to catalytic activity of the inducing protein, i.e., the nucleotides themselves, rather than growth-toxic effects of the induced protein. There is consensus that (p)ppGpp regulation of growth primarily reflects inhibition of total rRNA synthesis, exemplified by rrnB P1 promoter initiation activity. Systematic in vitro measurements on transcription initiation revealed that pure ppGpp is a more potent inhibitor of initiation than pppGpp, as predicted from growth and RNA/DNA ratio changes. Conversely, the activation of threonine promoter transcription initiation by ppGpp revealed it to be a more potent than pppGpp. Finally, ppGpp was again found to be stronger than pppGpp as an activator the gene expression changes mediated by RpoS, an alternative sigma factor responsible for protective adaptation when cells stop growing as nutrients are exhausted.
In all, potent regulation by ppGpp was always found to be shared to a lesser extent by pppGpp, with no examples of regulation unique to one or the other. This suggests that a site on RNA polymerase might bind to both ppGpp and pppGpp. Katsuhiro Murakami found the single site (1) that binds to both ppGpp and pppGpp, when crystals of RNA polymerase were soaked with each nucleotide. Two other groups reported a similar site for ppGpp within a few days of our publication. They also provided biochemical evidence that changing key residues does alter ppGpp binding kinetics by a variety of assays. The question of regulatory relevance has not been solved because the binding site is about 100 Å from the catalytic center. We will address this issue by determining whether regulation of mutant cells is rendered insensitive to (p)ppGpp accumulation.
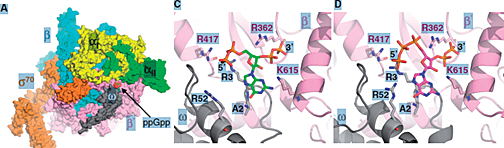
Figure 1. RNA polymerase subunits with ppGpp binding site (A), close-up view of binding of ppGpp (C) or pppGpp (D)
RpoC:pink; omega: grey
The search for new (p)ppGpp-like regulators
Direct identification of ppGpp in animal cells has proven difficult despite the high sensitivity of currently available chemical techniques, including mass-spectrometric metabolomic approaches. This is unexpected given that, in bacteria, ppGpp can reach mMolar levels, equal to GTP. This is in part attributable to the extreme metabolic lability of ppGpp (half-life about 20 seconds) coupled with its chemical instability under conditions normally used for isolating nucleotides. So far, (p)ppGpp has not been found in eukaryotes other than plants, and yet a functional, structurally bacteria-like ppGpp hydrolase (called Mesh-1) has been found to be biologically ubiquitous. If (p)ppGpp is indeed absent, we hypothesized that the Mesh1 hydrolases might function to degrade a compound similar to (p)ppGpp. We thus embarked on structure-specificity studies with purified Mesh1 proteins in search of possible candidates that could provide clues to substrates hydrolyzed by Mesh1.
In support of this idea, we found that classical animal Mesh hydrolases have extended substrate specificities compared with bacteria. Bacterial enzymes can remove the 3′-pyrophosphoryl groups of (p)ppGpp but not of (p)ppApp, the corresponding adenine analogs. In contrast, the human and fly Mesh hydrolases are able to cleave both guanine and adenine analogs. We plan to exploit this difference in substrate specificity in an attempt to deduce likely substrates in eukaryotes. We are taking two approaches. The first is based on bioinformatics: finding bacterial homologs of animal Mesh proteins in bacteria. We determine whether the substrate specificities of these enzymes are similar to those of animals and, if so, whether synthetases exist to generate appropriate substrates. The second approach is to compare existing protein structures of animal Mesh proteins with bacterial hydrolases to predict specificity determinants altered by mutations in the catalytic sites.
Possible extension of (p)ppGpp regulation to include bacterial viruses
There is growing evidence to suggest that (p)ppGpp-like regulation occurs in animals as well as some archea, bacteria, and plants (see above). We are taking advantage of the expertise gained by Tamara James’s doctoral work to explore the possibility that (p)ppGpp regulation might also occur during virus replication in bacteria. We studied the classical bacteriophage T4. Phage T4 development uses the E. coli–host transcriptional and translational machinery to produce about 100 mature virus particles that rapidly—20 minutes after infection—burst the cell. This can easily be viewed as extreme stress for the bacterial host. James studied phage mutants of MotA and AsiA proteins that function in this process to modify transcription so that promoters are activated to permit phage gene expression in later development. The proteins modify sigma factor remodeling so that RNA polymerase can acquire the ability to recognize phage T4 DNA sequences as promoters. She first determined whether E. coli mutants lacking DksA or (p)ppGpp alter phage T4 development and found that each mutant dramatically increases the size of clear phage plaques (3–4 times larger diameter), suggesting that the yield of infecting virus is enhanced when cellular stress defenses are compromised. Next, studies with phage mutants tested the possibility that the T4 phage small-plaque developmental phenotypes of asiA or motA mutants are affected by altered (p)ppGpp regulation. The tiny plaques of the phage mutants in wild-type hosts were found enlarged in both ppGppo and dksA mutant hosts, approaching wild-type plaque size. We are now focusing on details of the mechanisms. Current studies are determining whether premature promoter temporal activation is involved or whether promoter bypass occurs, i.e., whether activating each promoter individually or excessive activation of early promoters leads to read-through activation of both promoters. Either early event could amplify later T4 gene expression to enhance virus yield. The project is important because it promises to extend the range of regulatory effects of ppGpp and DksA to include bacterial virus gene transcription.
Regulatory effects of ppGpp on transcription
We seek to understand how ppGpp regulates gene expression through promoter-specific positive and negative regulatory effects on transcription. For years, we have focused on regulatory interactions between RNAP secondary channel–binding proteins (such as DksA, GreA, GreB) and ppGpp. Our recent analyses (2) confirm expectations from structural similarities and parallel channel binding to RNA polymerase that DksA and GreA/B can show similar and even redundant functions. One example is that GreA overproduction can override the amino acid requirements of the dksA mutant for growth in minimal medium (ILTV), which are a subset of ppGpp0 requirements (DEFHILSTV). A related example is that GreA or DksA overexpression can reverse some of the amino acid requirements shown by ppGpp0 cells. We used microarray transcription profiles to extend earlier findings of shared functions by GreA and DksA to the cellular transcriptome. This more detailed global analysis reveals instances of GreA and DksA acting in opposition. An example of opposite effects at the phenotypic level is that the complete reversal of the amino acid requirements of ppGpp0 cells by overexpression of GreA occurs only in the absence of DksA and vice versa. This surprising observation suggests that the presence of one protein is redundant to the effect of the other gene product but abolished by the simultaneous presence of both. This oddly symmetrical epistatic effect is reminiscent of synthetic gene lethals.
While redundancies exist, the effects of these gene products are not quantitatively equivalent. As judged by transcription profiling, more genes, i.e.,331, are affected by GreA overproduction when DksA is absent than when DksA is present, i.e., 45; yet genes affected by DksA when GreA is absent show a very modest skew. Profiling indicates that the strongest activation by GreA occurs for gadA and gadE genes. We could verify this with P gadA– and P gadE–lacZ promoter transcriptional fusions as reporters, which serve to again localize activation to the level of transcription initiation rather than elongation. To determine what functional attributes of GreA and DksA are responsible for their interactive effects on regulation, we used well known mutants of GreA and DksA with altered acidic residues in the tip of the coil-coil finger structure. When GreA or DksA is bound to RNA polymerase, the fingertip reaches through a secondary channel deep into RNA polymerase to reach a point near its catalytic center for transcription. There, the fingertip acidic residues are thought to provoke the classical effects of such factors on RNA chain elongation and relief of arrested elongation. While the mutants do abolish the classical functions, they do not alter the regulation of selected promoter–lacZ fusions. This surprising observation strongly suggests that the regulatory features we uncovered represent new phenomena not easily explained by known behavior.
These more detailed approaches reinforce our notions from earlier experiments that the effects of ppGpp involve a complex interplay between GreA, GreB, and DksA. Stated in another way, it can be argued that our experiments reveal a modulatory function for ppGpp for this interplay, which could be the primary cause of complex global changes in gene expression. Mechanistically, this could occur by promoter activation, competition for RNA polymerase at the level of either binding or after persistent secondary channel occupancy, as well as mutual control of the expression of other secondary channel proteins. Our transcriptional profiling studies serve to localize some of the interactions to specific genes and regulators, which might lead to new regulatory mechanisms at the molecular level. We also explored the interactions between the three factors in a hierarchical series of RNAP mutants of increasing severity that allow ppGpp0 strains to grow without amino acids. The work again confirms that the ppGpp dependence of factor interactions operates at the level of RNAP—unusual for bacteria, for which gene-specific regulation otherwise almost invariably involves proteins that recognize specific DNA sequences. Overall, our experiments provide evidence that ppGpp regulation operates through a more complex regulatory interplay between GreA, GreB, and DksA than has been appreciated.
Publications
- Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res 2013;41:6175-6189.
- Vinella D, Potrykus K, Murphy H, Cashel MJ. Effects on growth by changes of the balance between GreA, GreB and DksA suggest mutual competition and functional redundancy in Escherichia coli. Bacteriol 2012;194:261-273.
Collaborators
- James Kennison, PhD, Program in Genomics of Differentiation, NICHD, Bethesda, MD
- Katsuhiko S. Murakami, PhD, Center for RNA Molecular Biology, Pennsylvania State University, University Park, PA
Contact
For more information, email cashel@mail.nih.gov or visit smr.nichd.nih.gov.

