You are here: Home > Section on Vertebrate Development
Control of Ectodermal Development in Vertebrate Embryos

- Thomas D. Sargent, PhD, Head, Section on Vertebrate Development
- Hiu Wan Law, Visiting Fellow
- Yoko Ogawa, PhD, Visiting Fellow
- Valerie Virta, PhD, Postdoctoral Intramural Research Training Award Fellow
- Sebastian Bilitza, BA, Postbaccalaureate Fellow
This lab focuses on mechanisms regulating the differentiation of cranial neural crest cells that give rise to the bone and cartilage of the vertebrate jaw, gills, neurocranium and other structures of the head. Our approach is to manipulate cell-cell signaling and transcriptional control mechanisms in the intact zebrafish embryo using inducible transgenic strategies, aiming to identify the regulatory networks that control craniofacial development. Disruption of these developmental programs is the most common source of birth defects in humans, the detection, prevention, and treatment of which is a central aspect of the NICHD mission. Equally important, understanding the regulation of gene expression in this complex embryonic tissue represents a challenging and fascinating problem in basic molecular and developmental biology.
The origin of the lab’s neural crest research was our discovery in 2003 that the transcription factor TFAP2a is both necessary and sufficient to trigger the conversion of cells at the neural plate border from neural to neural crest identity in the frog Xenopus. We went on to show that TFAP2a mediates the transcriptional response to bone morphogenetic protein (BMP) signaling in neural crest induction and then identified several regulatory targets of TFAP2a using microarray analysis. The targets include the motor protein MyosinX, PCNS, a previously undiscovered protocadherin, and a completely novel gene we named Inka. The lab also carried out pioneering research on the homeodomain factor Dlx3, performing the first mouse knock-out of this gene and demonstrating its function in the development of mammalian epidermis and placenta. We also established the role for this factor in establishing the boundary of the neural crest in Xenopus.
The Inka project was considered high risk given that the gene is entirely novel with no homology to any known protein, but was attractive owing to the striking effects that overexpression of the gene had on cytoskeletal dynamics in both Xenopus embryos and in mammalian cell culture. We used antisense morpholino oligonucleotides (MOs) to “knock down” Inka expression in Xenopus and zebrafish embryos and found that the loss of function resulted in severe disruption of craniofacial development. However, in collaborations with Trevor Williams' lab and the Celia Moens' group, we succeeded in ablating the Inka gene in mouse and zebrafish, respectively and found—to our surprise and disappointment—that loss of Inka in both species failed to elicit a discernable phenotype. This suggested that the MO–generated phenotype might be misleading, and work on the Inka project has been put on hold until this issue is resolved.
The group has also investigated the function of pak4 (p21-activated kinase 4), a Rho-GTPase effector molecule that was identified earlier as an interaction partner for Inka. Experiments using MOs led to the conclusion that pak4 is a maternal-effect gene required for multiple aspects of organogenesis, including formation of myeloid cells (macrophages and granulocytes) and morphogenesis of axial muscles. Recently the group generated lines of zebrafish in which pak4 is ablated using TALEN-based gene targeting. Unexpectedly, homozygous pak4 null fish, even after three generations of inbreeding, appear to be normal and fertile. This suggests that, unlike in mammals, where pak4 is essential for early development, the gene may be dispensable in zebrafish. Furthermore, these results, in conjunction with those from the Inka loss-of-function experiments, lead us to conclude that even with extensive controls, use of MOs as a primary tool for establishing gene function can give misleading results and should probably not be relied upon as a tool for primary investigation of gene function, at least in zebrafish. We are working to understand the discrepancy between MO and genetic loss-of-function phenotypes to complete the pak4 project. Meanwhile, current research is to learn more about the molecular function of Dlx and TFAP2 factors and BMP signaling in neural crest, as outlined in the next sections.
Pak4 function in zebrafish
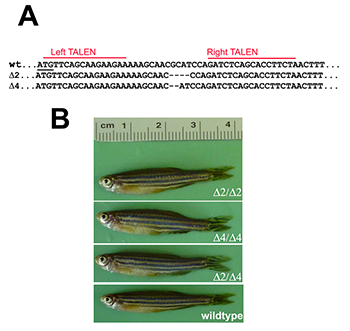
TALEN-mediated targeting of the single zebrafish pak4 gene yielded several INDEL mutations near the predicted cleavage site 20 bp downstream from the translational start methionine codon. Two of these, a two-base deletion (Δ2) and four-base deletion (Δ4) were chosen for further analysis (Figure 1A). As both should result in frameshifts eliminating virtually the entire pak4 protein, compound heterozygotes would be expected to be pak4 nulls. This was confirmed by Western blotting, using antisera we had raised against peptides located in a highly variable region in the protein. Our earlier work using MOs (see above) suggested that pak4 behaves as a maternal effect gene, so we incrossed mutant F2 fish, anticipating abnormal development in F3 embryos. Instead we observed completely normal development of healthy, fertile adults (Figure 1B). The most likely interpretation is that pak4 is not essential in zebrafish, and that our results indicating otherwise were attributable to an complex artifact tied to the MOs that were used in the loss-of-function study. However, our anti-pak4 antibody was targeted to a site located upstream of the kinase domain, so it is conceivable that somehow the mutant mRNAs were translated from internal ATG codons, yielding a partially functional protein that would be undetectable in Western blots. To test this hypothesis, we are making pak4-GFP fusion constructs with the full 5′ untranslated region, determined by the RACE method, linked to the full coding sequence with either wild type, Δ2, or Δ4 configurations at the mutation site. RNA and DNA injections will be carried out using GFP fluorescence as a read-out. If the embryonic translational machinery can somehow skip the normal ATG initiation codon, both mutant and wild-type constructs should express GFP; if not, the TALEN mutants presumably represent genuine nulls, and normal development is the correct loss-of-function phenotype.
Figure 1. Targeted disruption of the pak4 gene in zebrafish
TALEN nucleases were designed to recognize two 16–bp targets (A, red lines) located immediately downstream of the translation start codon (underlined). Several INDEL mutations were identified, including two deletions shown here that remove 2 and 4 base pairs, respectively. By crossing F1 heterozygous fish, we obtained F2 and then F3 generations that were homozygous for both mutations; we also obtained compound heterozygotes. Panel B shows selected individuals, none of which have any discernible abnormalities.
Dlx gene function in cranial neural crest development
Click image to enlarge.
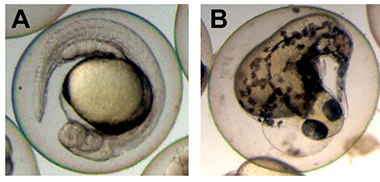
Figure 2. Disrupted development in embryos injected with engrailed-dlx3b RNA
Normal control (A) and abnormal injected (B) embryos are shown at 24 hours post fertilization. Expression of the dominant negative dlx3 construct resulted in multiple morphological defects that depended on the addition of the inducer TAM to the medium. This demonstrates the biological activity of and regulation by TAM induction.
In the past year, we set out to use transgenic zebrafish to test the theory that the mutated human DLX3 gene (DLX3TDO allele) acted as a dominant negative inhibitor of wild-type DLX3, accounting for the dominant inheritance of Tricho-Dento-Osseous syndrome (TDO). We also planned to use this mutated allele as a tool to disrupt NC development experimentally in zebrafish. To circumvent early functions of Dlx genes, and for added flexibility, we decided on a binary transgenic strategy, by which the zebrafish sox10 promoter drives expression of an ecdysone-inducible Gal4 activator, which could then activate expression of the DLX3TDO allele, residing in a separate transgene, at the desired time in the NC domain. We found that such expression did not significantly alter craniofacial development. However, the level of transgene expression was weaker than expected and never approached that of the endogenous dlx3 gene in the fish. This made it impossible to confirm or rule out the dominant negative model and also limited the utility of the tg lines. We then switched to an alternate strategy that may be less versatile, but one that we considered more likely to yield effects on the embryo. In this approach, the potent repressor domain from the Drosophila engrailed gene is fused to the extended homeodomain of the zebrafish dlx3b gene, followed by a modified estrogen receptor ligand binding domain (ERT2), which confers functional induction following treatment with the estrogen analog tamoxifen (TAM). Preliminary results with RNA injection show severe embryonic defects upon TAM treatment, indicating that the recombinant protein is biologically active and properly regulated (Figure 2). The functional cassette has been inserted into our sox10-driven transgenesis vector, and transgenic lines are being established. The plan is to use embryos from these lines to block Dlx gene function specifically in the NC (via the sox10 promoter) at specific times (via TAM induction). Of particular interest is to investigate Dlx function late in cranial NC development, after migration has been completed and when cranial bones, jaws, and other components begin to differentiate. This would add significantly to existing models of Dlx function based on gene targeting in mouse and knock down experiments in zebrafish, that latter being based on MO injection, which does not allow time-specific control and, as noted above, would benefit from corroboration using different methodology.
The longer-term goal of these experiments is to identify potential targets for Dlx gene regulation. RNA will be isolated from affected and control embryos and analyzed by our collaborator Maria Morasso’s laboratory for alterations in gene expression using RNA-seq technology provided by the NIAMS genomics core facility. Candidate genes will be screened by in situ hybridization to identify those with appropriate expression patterns in fish embryos. This will be followed by functional studies using gene targeting in zebrafish, and in suitable cases, in the mouse by Morasso’s group.
Role of BMP signaling in patterning craniofacial morphology
Our lab has a long-standing interest in signaling via BMP factors in ectodermal development. We were the first to show, in 1989, that dissociated Xenopus ectoderm switched from an epidermal to a neural fate. This was later shown to be the result of disruption of BMP signaling (and subsequently re-interpreted as a more fundamental effect on MAP kinase activation). Our work on TFAP2 showed that the factor can mediate the BMP–dependent aspects of NC induction in Xenopus and implicated the differential control of Dlx factors by a BMP activity gradient in setting up the NC boundary. In our current studies, we are using inducible expression of BMP ligands and dominant-negative BMP receptors in zebrafish NC to study the effects of changed BMP signaling at different times in development on the resulting cranial skeleton. We anticipate that this will provide a good experimental platform for further analysis on craniofacial patterning by BMP and potentially other ligand/receptor arrays.
TFAP2 function in cranial neural crest development
The project builds on the lab’s earlier work on the transcriptional activator protein TFAP2a, showing that the factor transmits BMP signaling to the gene-regulatory network and is necessary and sufficient for NC induction from neutralized ectoderm. Missing from the earlier studies, carried out in Xenopus, was manipulation of the time of expression: TFAP2a function was both ablated and restored from the outset of development. There was also no way at that time, in Xenopus or zebrafish embryos, to resolve the cell-autonomous functions of TFAP2, i.e., in the NC, from secondary effects of TFAP2 activity in adjacent tissues, primarily epidermis.
To address these issues, and to continue the identification of downstream regulatory targets of TFAP2 in the NC, we are using a dominant negative transgenic approach that parallels that described above for Dlx genes. In this case, the Engrailed repressor has been linked to a Xenopus TFAP2 from which the activation domain has been deleted (ΔAD). All other molecular components are the same, as is the general experimental strategy. The basic repressor construct has been tested in zebrafish embryos by RNA injection in preliminary experiments and found to be biologically active (Figure 2). The next steps will be to confirm induction control by TAM, produce transgenic lines, and identify and validate target genes by RNA-seq analysis, as outlined in the Dlx section above.
Publications
- Law HW, Sargent TD. Maternal expression of Pak4 is required for primitive myelopoiesis in zebrafish. Mech Dev 2013;130:181-194.
Collaborators
- Celia Moens, PhD, Fred Hutchinson Cancer Research Center, Seattle, WA
- Maria Morasso, PhD, Laboratory of Skin Biology, NIAMS, Bethesda, MD
- Trevor Williams, PhD, University of Colorado School of Medicine, Denver, CO