You are here: Home > Unit on Mammalian Epigenome Reprogramming
Epigenome Reprogramming During Mammalian Development

- Todd S. Macfarlan, PhD, Head, Unit on Mammalian Epigenome Reprogramming
- Carson Miller, PhD, Postdoctoral Fellow
- Gernot Wolf, PhD, Visiting Fellow
- Peng Yang, PhD, Visiting Fellow
- Warren Wu, BS, Postbaccalaureate Fellow
The overall goal of the laboratory is to understand how the epigenome is regulated in mammals during embryo development and to apply this knowledge to engineer tissues and build better models of disease. We explore how the genetic material is reprogrammed after fertilization to generate embryonic cells capable of generating all cell types that make up and support the embryo and how these pluripotent cells differentiate into specialized cell types. We utilize mouse genetics, developmental biology, biochemistry, cell biology, multi-dimensional imaging, stem cell–based in vitro modeling, and next-generation sequencing approaches to explore the role of histone-modifying enzymes and transcription factors during the reprogramming process.
Exploring the function of histone/DNA–modifying enzymes during development
A large number of enzymes that post-translationally modify histones/DNA have been identified and biochemically characterized. Many of these enzymes have also been shown to be essential for embryo development, mutated in cancers and neurological disease, or important for trans-generational epigenetic inheritance in model organisms. We are using stem cell–based models of development along with mouse genetics to remove enzymes that post-translationally modify histones and DNA in a tissue-specific manner. In particular, we are focusing on the period of preimplantation development, when massive reprogramming of the epigenome takes place, and during formation of the central nervous system, when cells are becoming post-mitotic as they differentiate into neurons and are thus incapable of replication-dependent changes in the epigenome. With these studies, we hope to gain a further understanding of the importance of epigenetic information stored in chromatin.
Interplay of transcription factors and chromatin-modifying enzymes
Transcription factors have long been known to be the master regulators of cell fate, binding to specific sequences of DNA and activating or silencing genes that allow specific cell types to carry out specialized functions. More recently, it has been demonstrated that transcription factors can convert somatic cells into induced pluripotent cells (iPS cells) or from one somatic type to another, although this process is relatively inefficient. One likely reason for the inefficiency is that differentiated cell types carry “restrictive” epigenomes that are less accessible to transcription factors than the “permissive” epigenomes of embryonic stem cells, which are easily reprogrammed to somatic cells. We are thus exploring the possibility that chromatin-modifying enzymes gate the activity of transcription factors during these reprogramming processes (both natural and artificial reprogramming), and we are testing how chromatin-modifying enzymes work in conjunction with transcription factors to regulate cell fate.
Exploring the regulation and function of endogenous retroviruses
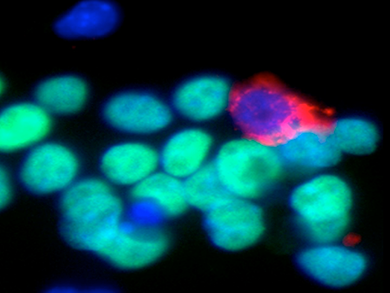
Figure 1. Activation of MERVL retroviruses within embryonic stem cell cultures
We mapped the transcriptome of two-cell (2C)–stage embryos by RNA sequencing and found that a large fraction of mRNAs are derived from endogenous retroviruses (ERVs), including several non-viral genes that utilize the ERV–LTRs (long terminal repeats) as their promoters. Using a reporter gene controlled by the LTR in ES cells, we identified a rare population of cells that had high LTR activity (labeled red in the accompanying image with antibodies against MERVL Gag proteins) and expressed genes thought to be restricted to the 2C stage of embryo development when the blastomeres are totipotent (as opposed to the pluripotent inner cell mass from which ES cells are derived). Using a Cre-Loxp–mediated fate-mapping strategy, we showed that all cells within ES cell cultures enter into this privileged state, whereby the cells take on totipotent-like characteristics, with the remarkable capacity to contribute to both embryonic and extra-embryonic tissues in mouse chimeras. The studies explain the classic experiments demonstrating rare cells within ES cultures that contribute to trophectoderm and placental tissues in mouse chimeras; the studies also suggest that ERV–LTRs have been co-opted by the host to control embryonic potency during development. The results raise the exciting possibility that 2C–restricted genes controlled by ERV–LTRs are essential to embryonic reprogramming and perhaps explain why somatic cell nuclear transfer (SCNT) is a highly inefficient process.
It is well known that retroviruses pose a threat to human health by infection of somatic cells, but retroviruses have also been infecting our mammalian ancestors for millions of years and have been incorporated into the germ-line as endogenous retroviruses (ERVs) that account for nearly 10% of our genomic DNA. We are interested in studying ERVs from two perspectives: as a parasite that must be kept in check by the host to prevent widespread viral activation and genomic instability; and as a symbiont that can be co-opted and utilized by the host for developmental and evolutionary advantage. Specifically, our work aims to study how the host has adapted molecular recognition machinery to transcriptionally silence ERVs, how the ERVs have sometimes evaded these silencing mechanisms during development, and how these evasive activities have led to host co-option of viral regulatory sequences that may have contributed to early cell fate choices in the pre-implantation embryos of placental mammals. The studies will lead to important insights into the molecular “arms race” between retroviruses and their hosts, which continue play out today, and will also lead to a better understanding of ERVs' integral role in driving our development and evolution as a species.
Publications
- Macfarlan TS, Gifford WD, Driscoll S, Rowe HM, Bonanomi D, Lettieri K, Firth A, Singer O, Trono D, Pfaff SL. ES cell potency fluctuates with endogenous retrovirus activity. Nature 2012;487:57-63.
- Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol 2013;218-226.
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 2013;23:452-461.
Collaborators
- Kai Ge, PhD, Laboratory of Endocrinology and Receptor Biology, NIDDK, Bethesda, MD
- Finn Skou Pedersen, PhD, Aarhus University, Aarhus, Denmark
- Samuel Pfaff, PhD, The Salk Institute and the Howard Hughes Medical Institute, La Jolla, CA
- Che-Kun (James) Shen, PhD, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan
- Didier Trono, MD, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Contact
For more information, email lovep@mail.nih.gov.