You are here: Home > Section on Developmental Biology
Molecular Genetics of Embryogenesis in Xenopus and Zebrafish

- Igor B. Dawid, PhD, Head, Section on Developmental Biology
- Mi Ha Kim, PhD, Visiting Fellow
- Minho Won, PhD, Visiting Fellow
- Valeria Zarelli, PhD, Visiting Fellow
- Alison M. Heffer, PhD, Intramural Research Training Award Fellow
- Martha Rebbert, BS, Senior Technician
- John Gonzales, MA, MS, Technician
- Nathaniel A. Parker, BA, Postbaccalaureate Student
The laboratory uses the frog Xenopus laevis and the zebrafish Danio rerio as experimental systems in which to study molecular-genetic mechanisms of early vertebrate development. Recently, we focused on mechanisms of caudal body development, neural crest specification, pancreas development, and targeted gene disruption.
The role of the homeodomain factor Hhex in pancreas development
One of the factors that emerged from our screening efforts is vvp1. In a collaborative effort, we found that the vvp1 gene is an excellent marker for a population of early ventral pancreatic precursors (1). Using this novel marker gene, we studied pancreas development in Xenopus embryos, focusing on the regulatory function of the homeobox gene hhex. In tail bud embryos, vpp1 expression can be used to specifically label two ventral pancreatic buds, whereas hhex expression is primarily seen in the liver diverticulum. Ectopic over-expression of a carefully titrated level of hhex mRNA led to a substantial increase in the domain of vpp1-positive cells, which later led to increased development of the ventral pancreas. These giant ventral pancreata may arise by a re-specification of intestinal precursor cells to ventral pancreatic precursor cells. In using antisense morpholino oligonucleotides to study loss-of-function phenotypes, the knockdown of hhex led to a reduction of the expression of the vpp1 gene and the specific interference with the development of the ventral pancreas. On the basis of these observations, we suggest that hhex plays an essential role in the formation of a vpp1-positive cell population in the endoderm of the gastrula embryo, a population that is essential for the subsequent development of the ventral pancreas.
Targeted gene disruption in Xenopus using TALENs
For a long time, the frog Xenopus has been a valuable model system in developmental biology. However, as in the zebrafish, the lack of techniques for targeted gene disruption in this model has been a major limitation to progress. Recent developments are changing this situation. Transcription activator-like effectors (TALEs), discovered in plant pathogenic bacteria, are DNA-binding proteins with a simple code that has been deciphered, allowing the generation of DNA-binding modules for any target sequence. TALE-nuclease hybrid proteins (TALENs) are able to cut the chromosome within the nucleus at the targeted site, leading to repair that often introduces deletions or insertions, disrupting the gene. In a collaborative effort, we applied and adapted the TALEN technology to Xenopus and showed that it functions at high efficiency in this system (2). Beyond the Xenopus system, we are in the process of further developing applications of the technology in the zebrafish.
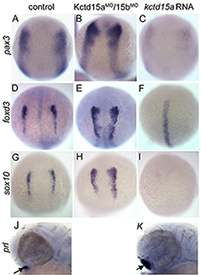
Figure 1. Kctd15 inhibits neural crest specification in zebrafish.
In situ hybridization of early somite embryos shows that markers of neural plate border (pax3) and neural crest (foxd3, sox10) are inhibited by Kctd15 overexpression and enhanced by its knock-down. Adapted from Dutta S, Dawid IB. Development 2010;137:3013-3018.
The BTB domain protein Kctd15 restricts the neural crest domain in zebrafish embryos and inhibits transcription factor AP-2.
Our laboratory has a long-standing interest in the formation of the neural crest (NC), a group of cells with stem cell properties, which arise at the dorsal neural tube and migrate to many locations in the embryos to give rise to a large number of varied differentiated derivatives. A recent focus has been the role of the BTB domain–containing protein Kctd15 in regulating the NC domain in the embryo. Kctd15 is first expressed at the neural plate border, where the NC and the pre-placodal domains are specified. Overexpression of Kctd15 strongly inhibits NC specification, whereas knock-down of Kcdt15 by antisense morpholinos enhances the expression of NC marker genes (Figure 1). We propose that Kctd15 is involved in delineating the size of the NC domain at the neural plate border.
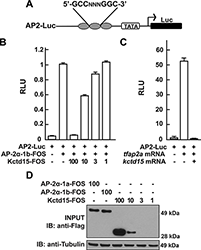
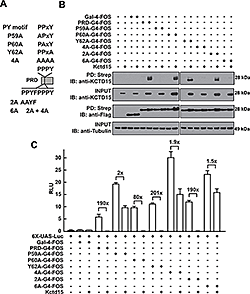
In pursuing the molecular mechanism of Kctd15 action, we found that Kctd15 regulates the activity of transcription factor AP-2. AP-2 is known to be a key factor in the induction and differentiation of NC cells. We found that AP-2 and Kctd15 can interact when co-expressed in cultured cells. Further, Kctd15 is a highly effective inhibitor of AP-2 activity as assayed using a reporter construct in cultured cells or in zebrafish embryos (Figure 2). In studying the mechanism of inhibition of AP-2 by Kctd15m we found that Kctd15 interacts with the activation domain of AP-2. We further analyzed this system using a fusion product between the Gal4 DNA–binding domain and the AP-2–activation domain. Within the latter domain, a conserved proline-rich motif proved critical for Kctd15 interaction: mutation of proline 59 to alanine (P59A) in the Gal4-AP-2 fusion resulted in a protein that was active but could not bind to and was insensitive to inhibition by Kctd15 (Figure 3). This proline residue was also essential for Kctd15 sensitivity in the context of the full-length AP-2 molecule. Thus, we conclude that Kctd15 inhibits AP-2 by binding to a specific site in its activation domain (3).
Figure 2. Kcdt15 represses AP-2 function.
(A) AP2-Luc reporter, containing three AP-2 consensus sites driving expression of luciferase (Luc). (B) The reporter was strongly stimulated by zebrafish AP-2α and dramatically inhibited by zebrafish Kctd15 (levels in ng indicated). RLU, relative light units. (C) Reporter activity in zebrafish embryos. (D) Cells were transfected with FOS-tagged zebrafish AP-2α and Kctd15, and lysates were blotted to assay expression of both proteins. From Zarelli & David, 2013 (3).
In additional functional studies, we determined that Kctd15 can be SUMOylated at a site close to its C terminus. SUMOylation is often associated with transcriptional repression, but we found that inhibition of AP-2 does not require SUMOylation of Kctd15. We speculate that Kctd15 carries out other biological roles that may depend on its ability to be SUMOylated (4).
Figure 3. Critical role of P59 for Kctd15 interaction and inhibition of AP-2
(A) Mutants in the conserved PPxY motif within the AP-2α proline-rich domain (PRD) and two additional mutants in the adjoining region are shown. PRDs were fused to the Gal4-DNA binding domain and the FOS tag. (B) Kctd15 interaction depends on P59. WT and mutant fusion constructs were expressed in HEK293T cells alone or with Kctd15, and complexes were pulled down using a Strep-Tactin matrix. PRDs could bind to Kctd15 except in the constructs containing the P59A mutation (P59A, 4A and 6A). (C) PPxY-mutants activate the UAS-Luc reporter two-to-five fold more highly than WT PRD-Gal4. PRD-Gal4 WT and P60A, Y62A and 2A were dramatically inhibited by zebrafish Kctd15, whereas P59A and mutants containing this change were inhibited two-fold or less. The constructs were expressed at similar levels (B). Figure from Zarelli & David, 2013 (3).
Habenular commissure formation in zebrafish is regulated by the pineal gland–specific gene unc119c.
Click image to enlarge.
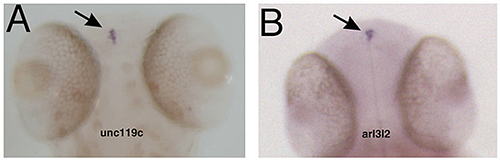
Figure 4. Specific gene expression in the pineal
In situ hybridization showing that unc119c and arl3l2 are specifically expressed in the pineal gland. Adapted from Toyama et al. Dev Dyn 2009;238:1813–1826.
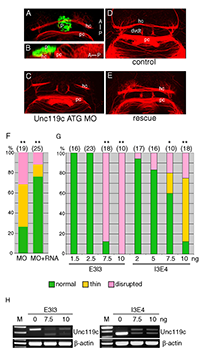
We have studied the pineal gland, a brain region critical for the control of circadian rhythm, by characterizing the pineal transcriptome at different stages of zebrafish development. Functional analysis of an individual gene with differential expression in the pineal gland, named unc119c, was completed, showing that this gene is required for the habenular commissure to extend across the midline (5). The Unc119 family is represented by two genes in the human, but the gene we isolated is the third family member noted in zebrafish. This gene, which we named unc119c, is specifically expressed in the pineal, with a low level of expression in the retina (Figure 4). In fish the pineal is a photosensitive organ and shows many similarities in gene expression to the retina. Unc119 proteins interact with small GTPases of the Arl3 family, and we showed that Unc119c binds to Arl3l2 when both are coexpressed in heterologous cells. Using morpholino antisense oligonucleotide (MO)–mediated knock-down of unc119c expression, we found that the protein is required for the formation of the habenular commissure (HC) in the zebrafish (Figure 5). The HC crosses the midline in close proximity to the pineal gland. Knock-down of the Unc119c binding partners Arl3l1 or Arl3l2 also affects HC formation. We hypothesized that Unc119c might be involved in protein trafficking or secretion of a guidance factor involved in HC formation. Based on the literature and expression pattern, we picked Wnt4a as a candidate target gene; Wnt4a has been reported by others to be required for HC formation, a fact confirmed in our experiments. We found that Wnt4a accumulation and secretion from cultured HEK293T cells is stimulated by Unc119c and Arl3l1/2. Thus, we propose that Unc119c is required in the pineal gland to stimulate Wnt4a secretion, while Wnt4a in turn acts as a guidance cue for the HC as it traverses the zebrafish forebrain (5).
Figure 5. Unc119c is involved in HC formation in zebrafish embryos.
(A, B) Visualizing the HC. Transgenic zebrafish, Tg(AANAT2:eGFP), which express eGFP in the pineal gland, were used. The embryos were fixed at 54 hpf and stained with anti-GFP (green, pineal gland) and anti-acetylated tubulin (red, axonal tracts). The HC is located directly beneath the pineal gland. Dorsal view (A), lateral view (B). (C-G) Injection of unc119c MO disrupts HC formation. Wild-type zebrafish embryos were injected with control MO (C), unc119c ATG MO (D), or unc119c ATG MO together with unc119c RNA (E, F). Two different unc119c splice MOs, E3I3 and I3E4, also affected HC formation in a dose-dependent manner (G). Statistical significance is indicated as * signifying p<0.05, and ** signifying p<0.01. Experiments are compared with HC in control embryos (which were all normal), except for the rescue data (F, second bar, MO+RNA), which are compared with MO-injected embryos (F, first bar).The splice MOs greatly reduced formation of mature mRNA (H). dvdt, dorsoventral diencephalic tract; hc, habenular commissure; p, pineal gland; pc, posterior commissure. From Toyama et al., 2013 (5).
Additional Funding
- NICHD, DIR
Publications
- Zhao H, Han D, Dawid IB, Pieler T, Chen Y. Homeoprotein hhex-induced conversion of intestinal to ventral pancreatic precursors results in the formation of giant pancreata in Xenopus embryos. Proc Natl Acad Sci USA 2012;109:8594-8599.
- Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Ki H, Cheng C, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc Natl Acad Sci USA 2012;109:17484-17489.
- Zarelli VE, Dawid IB. Inhibition of neural crest formation by Kctd15 involves regulation of transcription factor AP-2. Proc Natl Acad Sci USA 2013;110:2870-2875.
- Zarelli VE, Dawid IB. The BTB-containing protein Kctd15 is SUMOylated in vivo. PLoS One 2013;8(9):e75016.
- Toyama R, Kim MH, Rebbert ML, Gonzales J, Burgess H, Dawid IB. Habenular commissure formation in zebrafish is regulated by the pineal gland-specific gene unc119c. Dev Dyn 2013;242:1033–1042.
Collaborators
- Harold Burgess, PhD, Program in Genomics of Differentiation, NICHD, Bethesda, MD
- Hui Zhao, PhD, The Chinese University, Hong Kong, China
Contact
For more information, email dawidi@mail.nih.gov or visit sdb.nichd.nih.gov.