You are here: Home > Section on Human Genetics
The Chromatin-Based Epigenome in Development and Aging

- Bruce H. Howard, MD, Head, Section on Human Genetics
- Valya Russanova, PhD, Staff Scientist
- Irene Stevens, BS, Postbaccalaureate Intramural Research Training Award Fellow
The normal human lifespan is marked by a complex series of developmental transitions, with relative stability during adulthood and, ultimately, a gradual decline in viability. Clock-like mechanisms presumably underlie the developmental events that occur through childhood and adolescence. Further, instabilities in such mechanisms are likely to be an integral part of the aging process and to contribute to many common degenerative diseases of later life. A breakthrough in the area of developmental transitions would have remarkable implications, and our research in this area reflects today's widespread interest in the rapidly evolving field of epigenomics.
Whole-genome surveys of epigenome structure
The past year saw continued application of next-Gen sequencing approaches to studies that use human tissues from several clinical sources. The sources include human skin fibroblasts from newborns and adults, as well as (through collaborative work with Alan DeCherney and James Segars), human ovarian granulosa cells harvested in association with ART (assisted reproductive technology) services at Shady Grove Hospital (Gaithersburg, MD).
With both experimental systems, we investigate developmental and age-related changes in chromatin structure, DNA methylation, and gene expression. In mammalian cells, cis-dependent epigenetic states are, of course, maintained by both chromatin structure and DNA methylation. We assess chromatin states with respect to histone acetylation and histone methylation patterns, as well as the topologies of these patterns extending over domains that typically encompass one or more genes. In recent experiments, we characterized DNA methylation at the whole genome level using both MethylCap-seq and Reduced Representation Bisulfite (RRBS). Of particular interest are interactions between the chromatin- (e.g., histone H3-K27 methylation) and DNA methylation–based components of epigenetic control.
Recent experiments with granulosa cells employed RRBS and RNA-seq to characterize methylation patterns and gene expression, respectively. Combining these and additional approaches, whole-genome surveys can be focused to explore epigenome features that underlie changes in gene function. Along these lines, bioinformatics analyses have revealed that: (i) transcribed regions with high content of CpG-enriched regions are predominantly down-regulated with age; (ii) down-regulation is associated with low levels of DNA methylation in such regions; and (iii) these highly significant trends are enhanced where such regions are localized near gene 3′ ends. Taken together, our findings suggest a previously unappreciated role for post-transcriptional mechanisms, e.g., nuclear processing or export, in controlling gene expression changes that accompany age-related decline in tissue function.
Given the progressive enhancements of high-throughput sequencing–based approaches in chromatin immunoprecipitation (ChIP-seq), RRBS, and RNA expression studies (RNA-seq), continuing refinement of bioinformatics tools is essential. We constantly improve and extend our genome annotation, pattern recognition, and pattern comparison algorithms. For topology-based mapping of chromatin patterns, we anticipate increasing use of the very deep datasets available through the NIH Roadmap Epigenomics and ENCODE programs. For this work, bioinformatic tools that efficiently link patterns in RNA expression data sets with epigenome features (see figure below) will be especially valuable.
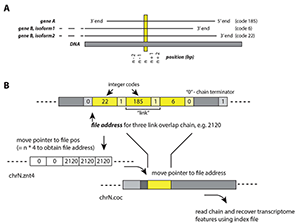
Figure 1. Virtually binary file–based genome annotation for overlapping features
A. An example of a genomic location (n) and its multiple associated features: gene A, gene B isoform 1, gene B isoform 2. Each feature is assigned an integer code (e.g., 185, 6, 22) in the concatenated annotation file.
B. The set of algorithms for locating n and recovering its associated features. Three files are required: annotation file (.znt4), concatenated file (.coc), and index file. The .znt4 and .coc files are opened in binary mode, and contain information in integer (4-byte units), and composite integer + character (5-byte units), respectively.
- Step 1. Move file pointer to chrN.znt4 file position (= n*4), to obtain address in chrN.coc file, e.g., 2120.
- Step 2. In chrN.coc file, move pointer to file address 2120, which is associated with a 3-link chain. Each link represents a feature. Links are separated by a “continue” integer “1”; each chain is terminated by an integer “0”.
- Step 3. Using index file, the integer codes recovered from Step 2 can be related to respective features.
Our chromatin-related results to date indicate that genes subject to both differentiation and developmental controls depend on the three-dimensional topology of the genome and are sensitive to the remodeling of higher-order chromatin structures. We hope to complete studies on large (on the order of 100 kb) domains over which histone acetylation or histone H3–K27 patterns are altered, as well as on control elements that may assume R loop or other non–B DNA conformations. DNA methylation–related results indicate that a small set of regions with high homology—either within gene clusters or dispersed gene families—may be substantially enriched for post-natal developmental- and age-related epigenome remodeling. Likewise, regions characterized by random monoallelic DNA methylation may be of special interest. If these and other related hypotheses are correct, new lines of investigation will be opened into the nature of clock-like epigenome functions.
The long-term goal for this field will be to generalize paradigms of dynamic postnatal epigenome structure so as to address, in innovative ways, a range of current problems in maternal reproductive health, pediatrics, and age-related disease.
Publications
- Salpea P, Russanova V, Hirai T, Sourlingas TG, Sekeri-Pataryas KE, Romero R, Epstein J, Howard BH. Postnatal development- and age-related changes in DNA methylation patterns in the human genome. Nucleic Acids Res 2012;40:6477-6494.
- Cole HA, Howard BH, Clark DJ. Genome-wide mapping of nucleosomes in yeast using paired-end sequencing. Methods Enzymol 2012;513:145-168.
- Nagarajavel V, Iben JR, Howard BH, Maraia RJ, Clark, DJ. Global 'bootprinting' reveals the elastic architecture of the yeast TFIIIB-TFIIIC transcription complex in vivo. Nucleic Acids Res 2013;41:8135-8143.
Collaborators
- Alan DeCherney, MD, Program in Reproductive and Adult Endocrinology, NICHD, Bethesda, MD
- Jonathan Epstein, MS, Molecular Genomics Laboratory, NICHD, Bethesda, MD
- James Segars, MD, Program in Reproductive and Adult Endocrinology, NICHD, Bethesda, MD
Contact
For more information, email howard@helix.nih.gov.