You are here: Home > Unit on Mammalian Epigenome Reprogramming
Epigenome Reprogramming During Mammalian Development

- Todd S. Macfarlan, PhD, Head, Unit on Mammalian Epigenome Reprogramming
- Carson Miller, PhD, Postdoctoral Fellow
- Gernot Wolf, PhD, Visiting Fellow
- Peng Yang, PhD, Visiting Fellow
- Don Hoang, BS, Postbaccalaureate Fellow
- Matthew Tinkham, BS, Postbaccalaureate Fellow
- Warren Wu, BS, Postbaccalaureate Fellow
- Sherry Ralls, BA, Biologist
- Justin Demmerle, AB, Graduate Student
The overall goal of the laboratory is to understand how the epigenome is regulated in mammals during embryo development and to apply this knowledge to engineer tissues and build improved models of disease. We explore how the genetic material is reprogrammed after fertilization to produce embryonic cells capable of generating all cell types that make up and support the embryo and how these pluripotent cells differentiate into specialized cell types. We utilize mouse genetics, developmental biology, biochemistry, cell biology, multi-dimensional imaging, stem cell–based in vitro modeling, and next-generation sequencing approaches to explore the interplay between histone-modifying enzymes and transcription factors during the reprogramming process.
Regulation of endogenous retroviruses by KRAB-zinc finger proteins
Click image to enlarge.
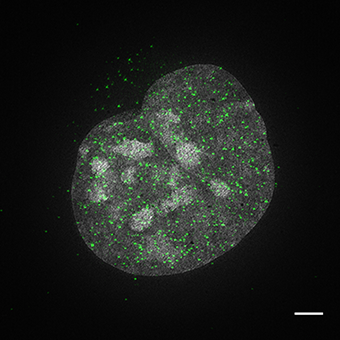
Nuclear distribution of ZFP809 in mouse ES cells
Flag-tagged ZFP809 was expressed in embryonic stem cells. ES cells were fixed and stained with anti-Zfp809 antibodies using indirect immunofluorescence (labeled in green). Cells were counterstained with DAPI and imaged using 3D Structured Illumination Microscopy (SIM). The image represents an individual nucleus with Zfp809 speckles found throughout the nucleus. 3D-SIM imaging was performed on a DeltaVision OMX V4 Blaze platform (GE Healthcare) with a 60X 1.42 NA PlanApo oil immersion objective (Olympus); 405- and 488-nm diode lasers; and PCO.Edge sCMOS cameras. Raw data were acquired with optical section of 125 nm and 15 raw SI images per plane per channel (five phases, three angles). Raw SI datasets were computationally reconstructed with softWoRx 6.0 software to obtain a super-resolution 3D image stack, and channels were aligned in the same program. The reconstructed image was manipulated in FIJI software to remove negative values from the 32-bit image, then converted to 8-bit where brightness and contrast were adjusted for presentation before creating a Z maximum projection.
It is well known that retroviruses pose a threat to human health by infecting somatic cells, but retroviruses have also been infecting our mammalian ancestors for millions of years and have been incorporated into the germ-line as endogenous retroviruses (ERVs), which account for nearly 10% of our genomic DNA. We are interested in studying ERVs from two perspectives: as a parasite that must be kept in check by the host to prevent widespread viral activation and genomic instability; and as a symbiont that can be co-opted and utilized by the host to developmental and evolutionary advantage. Specifically, our work aims to study how the host has adapted molecular recognition machinery to transcriptionally silence ERVs, how the ERVs have sometimes evaded the silencing mechanisms during development, and how the evasive activities have led to host co-option of viral regulatory sequences that may have contributed to early cell-fate choices in the pre-implantation embryos of placental mammals. The studies will lead to important insights into the molecular “arms race” between retroviruses and their hosts, which continue play out today, and will also lead to a better understanding of ERVs' integral role in driving our development and evolution as a species.
Kruppel-associated box zinc finger proteins (KRAB-ZFPs) are transcriptional repressors that make up the largest family of transcription factors in mammals (more than 500 members in mice and humans). KRAB-ZFPs also evolve rapidly. But strikingly little is known about the physiological functions of the majority of family members. The proteins consist of an N-terminal KRAB domain, which mediates recruitment of the co-repressor proteins KAP1/Trim28 and the histone H3K9 methyltransferase SETDB1, and a variable number of C-terminal zinc-finger domains that mediate sequence-specific DNA binding. We and others have hypothesized that KRAB-ZFPs function as a type of slowly adaptive immune system that transcriptionally silences ERVs. Several lines of evidence point to a potential function of the KRAB-ZFP family in binding to and silencing of ERVs. First, the number of KRAB-ZFPs in mammals tightly correlates with the number of ERVs and closely related LTR transposons. Second, the KRAB-ZFP family member ZFP809 was isolated based on its ability to bind to the primer-binding site for proline tRNA (PBS-Pro) of murine leukemia virus (MuLV). Third, deletion of the KRAB-ZFP co-repressor KAP1 leads to transcriptional activation of many families of ERVs in mouse ES cells. We are therefore testing whether KRAB-ZFPs are ERV silencers in the mouse.
To begin our analysis of KRAB-ZFPs, we focused on the function of the KRAB-ZFP family member ZFP809, which has previously been linked the silencing of exogenous MuLVs. Using ChIP-seq, mouse knockouts, epigenome profiling, and evolutionary analysis, we demonstrated that ZFP809 functions to silence ERVs by binding to an 18 nt motif corresponding to the primer binding site for proline (PBS-Pro). We showed that some of these ERVs are reactivated in Zfp809 knockout (KO) tissues, with a corresponding shift from repressive to active histone marks.
Several lines of evidence support the view that the critical window of ZFP809 function occurs around the time of implantation. First, ZFP809 binds to ERVs in ES cells. Second, ZFP809 protein levels are considerably higher in ES cells than somatic cells. Third, phenotypes in Zfp809 KO primary mouse embryonic fibroblasts (PMEFs) cannot be rescued by re-expression of ZFP809. Fourth, conditional deletion of Zfp809 in PMEFs does not phenocopy the ERV reactivation phenotype in PMEFs derived from Zfp809 KO mice. These data suggests that Zfp809 is required to initiate, but not fully maintain, the stable heterochromatic silencing of ERVs during embryo development.
It has been suggested that, as an evolutionary adaptation to reduce genome instability, KRAB-ZFPs may evolve in response to endogenization of retroviral elements. We tested the hypothesis by searching muroidea for the presence of ZFP809 orthologs and Pro-PBS–containing target ERVs. We demonstrated that the appearance of ZFP809 in muroidea predates the appearance of ERVs presently targeted by ZFP809, suggesting that ZFP809 evolved its DNA–binding domain against invading (exogenous) retroviruses. Nonetheless, our data provide a compelling case for a KRAB-ZFP that functions rather specifically to transcriptionally silence retroviruses via the recruitment of heterochromatin machinery in a mammalian organism.
Additional Funding
- Scientific Director's Award, 2014-2015, Epigenetic Memory that directs iPSC reprogramming: the role of H3.3
Publications
- Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol 2013;218-226.
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 2013;23:452-461.
- Yang P, Wu W, Macfarlan TS. Maternal histone variants and their chaperones promote paternal genome activation and boost somatic cell reprogramming. Bioessays 2014;E-pub ahead of print.
Collaborators
- Finn Skou Pedersen, PhD, Aarhus University, Aarhus, Denmark
- Keiko Ozato, PhD, Program in Genomics of Differentiation, NICHD, Bethesda, MD
Contact
For more information, email macfarlants@mail.nih.gov.

