You are here: Home > Section on Vertebrate Development
Control of Ectodermal Development in Vertebrate Embryos

- Thomas D. Sargent, PhD, Head, Section on Vertebrate Development
- Hiu Wan Law, Visiting Fellow
- Yoko Ogawa, PhD, Visiting Fellow
- Valerie Virta, PhD, Postdoctoral Intramural Research Training Award Fellow
- Mariam Awad, BA, Postbaccalaureate Fellow
- Sebastian Bilitza, BA, Postbaccalaureate Fellow
The lab focuses on mechanisms regulating the differentiation of cranial neural crest cells that give rise to the bone and cartilage of the vertebrate jaw, neurocranium, and other structures of the head. Our approach is to manipulate transcriptional control mechanisms in the intact zebrafish embryo using inducible transgenic strategies, with the aim of identifying the regulatory networks that control craniofacial development. Disruption of these developmental programs is the most common source of birth defects in humans, the detection, prevention, and treatment of which is a central aspect of the NICHD mission. Equally important, understanding the regulation of gene expression in this complex embryonic tissue represents a challenging and fascinating problem in basic molecular and developmental biology.
Click image to enlarge.
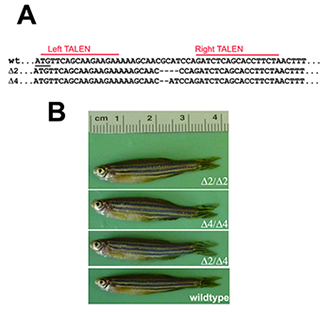
Figure 1. Targeted disruption of the pak4 gene in zebrafish
TALEN nucleases were designed to recognize two 16–bp targets (A, red lines) located immediately downstream of the translation start codon (underlined). Several INDEL mutations were identified, including two deletions shown here that remove 2 (Δ2) and 4 (Δ4) base pairs, respectively. By crossing F1 heterozygous fish, we obtained F2 and then F3 generations that were homozygous for both mutations; we also obtained compound heterozygotes. Panel B shows selected individuals, none of which have any discernible abnormalities.
The origin of the lab’s neural crest research was our discovery in 2003 that, in the frog Xenopus, the transcription factor TFAP2a is both necessary and sufficient to trigger the conversion of cells at the neural plate border from neural to neural crest identity. We went on to show that TFAP2a mediates the transcriptional response to bone morphogenetic protein (BMP) signaling in neural crest induction and then, using microarray analysis, identified several regulatory targets of TFAP2a, including a novel gene we called Inka. The lab also carried out pioneering research on the homeodomain factor Dlx3, performing the first mouse knock-out of this gene and demonstrating its function in the development of mammalian epidermis and placenta. We also investigated the role of the factor in establishing the boundary of the neural crest in Xenopus.
The Inka project was attractive owing to the striking effects that overexpression of the gene has on cytoskeletal dynamics in both Xenopus embryos and in mammalian cell culture. We used antisense morpholino oligonucleotides (MOs) to “knock down” Inka expression in Xenopus and zebrafish embryos and found that the loss of function resulted in severe disruption of craniofacial development. However, in collaborations with labs headed by Trevor Williams and Cecilia Moens, we succeeded in disrupting the Inka gene in mouse and zebrafish, respectively, but found that loss of Inka in both species failed to elicit a discernable phenotype. This suggested that the MO–generated phenotype was an artifact.
The group also investigated the function of pak4 (p21-activated kinase 4), a Rho–GTPase effector molecule that was identified earlier as an interaction partner for Inka. Experiments, again using MOs, led to the conclusion that pak4 is a maternal-effect gene required for multiple aspects of organogenesis, including formation of myeloid cells (macrophages and granulocytes) and morphogenesis of axial muscles. We were more confident of these results because of the extensive controls we carried out, including showing that the MO phenotype could be rescued by injecting pak4 mRNA lacking the MO binding site. However, we generated lines of zebrafish in which pak4 is ablated using TALEN–based gene targeting (Figure 1A). Unexpectedly, homozygous pak4 null fish, even after three generations of inbreeding, appear to be normal and fertile (Figure 1B), suggesting that, unlike in mammals, where pak4 is essential for early development, the gene is dispensable in zebrafish. Furthermore, the results, in conjunction with those from the Inka loss-of-function experiments, led us to conclude that even with extensive controls, use of MOs can give misleading results and should not be relied upon as a tool for primary investigation of gene function, at least in zebrafish.
Our current research focus is the molecular function of Dlx factors.
Dlx gene function in cranial neural crest development
We generated several zebrafish lines in which dominant negative versions of Dlx1 and Dlx3 were driven specifically in the neural crest (and also some placodal tissues) by inducible expression strategies based on the sox10 promoter. So far, we have not found any differences in craniofacial development in these lines compared with control zebrafish embryos. One possibility is that prior work on dlx genes in zebrafish suggesting important functions for these genes in jaw and other cranial bone/cartilage development was misleading, perhaps owing to artifacts associated with the morpholino oligonucleotides used in those experiments, and because dlx genes are not as important in zebrafish as they have been reported to be in mammalian facial development. However, it is also possible that our dominant negative constructs did not function as predicted or that expression levels were too low to overcome endogenous dlx factors. We are testing this by using a different transgenic approach based on a potent, inducible heat shock promoter, with neural crest specificity, provided by a cre/lox recombination system driven by the sox10 promoter. Production and testing of these lines are under way.
Publications
- Law SHW, Sargent TD. The serine-threonine protein kinase PAK4 is dispensable in zebrafish: identification of a morpholino-generated pseudophenotype. PLoS One 2014;E100268.
Collaborators
- Celia Moens, PhD, Fred Hutchinson Cancer Research Center, Seattle, WA
- Maria Morasso, PhD, Laboratory of Skin Biology, NIAMS, Bethesda, MD
- Trevor Williams, PhD, University of Colorado School of Medicine, Denver, CO

