You are here: Home > Section on Gene Expression
Control of Gene Expression during Development

- Judith A. Kassis, PhD, Head, Section on Gene Expression
- J. Lesley Brown, PhD, Staff Scientist
- Yuzhong Cheng, PhD, Senior Research Technician
- Sandip De, PhD, Postdoctoral Fellow
- Payal Ray, PhD, Postdoctoral Fellow
- Daniel Gutierrez, Summer Student
During development and differentiation, genes either become competent to be expressed or are stably silenced in an epigenetically heritable manner. This selective activation/repression of genes leads to the differentiation of tissue types. Recent evidence suggests that modifications of histones in chromatin contribute substantially to determining whether a gene is expressed. Our group is interested in understanding how chromatin-modifying protein complexes are recruited to DNA. In Drosophila, two groups of genes, the Polycomb group (PcG) and Trithorax group (TrxG), are important for inheritance of the silenced and active chromatin state, respectively. Regulatory elements called Polycomb group response elements (PRE) are cis-acting sequences required for the recruitment of chromatin-modifying PcG protein complexes. TrxG proteins may act through the same or overlapping cis-acting sequences. Our group aims to understand how PcG and TrxG proteins are recruited to DNA. We are also interested in how distantly located transcriptional enhancer elements selectively activate a promoter that may be tens (or even hundreds) of kilobases away. Our data suggest that promoter-proximal elements, some of which overlap with PREs, impart specificity to promoter-enhancer communication. We are also interested in the coordination between activation by enhancer elements and silencing by PcG proteins. Our data suggest that certain types of transcriptional activators may be able to overcome the repressive activity of PcG proteins. This may be particularly important at the target gene engrailed, given that PcG proteins are bound to engrailed PREs in both cells that repress and those that actively transcribe engrailed. Finally, our lab recently found that enhancing cohesin binding stability interferes with PcG repression.
Polycomb response elements
Click image to enlarge.
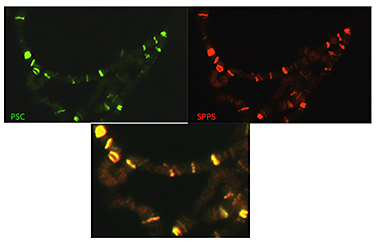
Figure 1. Spps and Psc co-localize on polytene chromosomes.
Polytene chromosomes were fixed and incubated with antibodies against Psc (green) and Spps (red). The lower panel shows that the two proteins completely co-localize on these chromosomes.
PREs are DNA elements that recruit Polycomb group (PcG) proteins to the DNA. PcG proteins act in protein complexes that repress gene expression by modifying chromatin. PcG protein complexes specifically associate with PREs in vivo; however, it is not known how they are recruited or held at the PRE. PREs are complex elements that harbor binding sites for many proteins. Our laboratory has been working to define all sequences and DNA-binding proteins required for the activity of a 181-bp PRE from the Drosophila gene encoding engrailed (en). At least seven DNA-binding sites contribute to the activity of this 181-bp PRE. One of the required binding sites is for the Polycomb-group protein Pleiohomeotic (Pho) and the related protein Pleiohomeotic-like (Phol). Binding sites for the proteins GAGA factor/Pipsqueak, Zeste, and Dsp1 are also present within the en PRE. Our laboratory found that members of the Sp1/KLF family of zinc-finger proteins bind to another required binding site. These proteins have undergone extensive study, revealing 20 Sp1/KLF family members in mammals. Drosophila has 9 Sp1/KLF family members, of which eight bind to the en PRE. We derived a consensus binding site for the Sp1/KLF Drosophila family members and showed that this consensus sequence is present in most molecularly characterized PREs. The data suggest that one or more Sp1/KLF family members play a role in PRE function in Drosophila. We identified one of the Sp1/KLF family members as being specifically associated with PREs. We named this protein Spps (Sp1 Factor for pairing-sensitive silencing). Our work shows that Spps binds to Drosophila polytene chromosomes in a pattern that completely overlaps with that of the PcG protein Psc (Figure 1). Chromatin immunoprecipitation experiments showed that Spps binds to PREs. The data strongly suggest that Spps plays a role in Polycomb-group repression. Using homologous recombination, we deleted the Spps gene and showed that it is required for Polycomb-group repression late in development. Finally, we showed that a mutation in Spps enhances the phenotype of Pho mutants, strongly suggesting that Spps and Pho work together to recruit Polycomb-group complexes to DNA (1). Thus, while Spps functions in PcG silencing, we do not know whether other Sp1/KLF family members also function in that manner. We made mutants in two other Sp1/KLF family members and are currently assessing their role, if any, in PcG function.
Although much is known about the protein-binding sites required for PRE function, we are not able to predict the location of a PRE based on the presence of binding sites alone. To help us identify either other protein-binding sites required for PRE function or other important characteristics of PREs (such as the number or spacing of binding sites), we have begun to analyze other PREs from the en region of the genome. To this end, we characterized two PREs from inv (which encodes invected), a gene that is co-regulated with en, and another PRE upstream of the en transcription start site. Our work suggests that, while binding sites for some proteins (Pho, Spps, GAGA factor) are required for the activity of all en/inv PREs, the arrangement, order, and number of binding site for each protein are variable in different PREs. We are in the process of determining the identity of other PRE-DNA–binding proteins.
The role of PREs and flanking sequences at the en gene
The Drosophila en gene encodes Engrailed, a homeodomain protein that plays an important role in the development of many parts of the embryo, including formation of the segments, nervous system, head, and gut. The gene also plays a particularly significant role in the development of the adult, specifying the posterior compartment of each imaginal disk. Accordingly, en is expressed in a highly specific and complex manner in the developing organism. We have been studying the 181–bp en PRE, which is located near the en promoter from −576 to −395 upstream of the transcription start site. We were interested in determining the role of this PRE in the control of en expression. One of our first findings demonstrated that this PRE is redundant with other flanking PREs in the endogenous en gene; another strong PRE is located from −1100 to −1500, and probably other weak PREs are located nearby. In fact, when we examined the location of Ph and Pho proteins on en DNA by chromatin immunoprecipitation (ChIP), we found that they are bound to a 2.5–kb region extending from the en promoter to about −2.5kb upstream. It is therefore perhaps not surprising that a 500–bp deletion that includes the 181–bp PRE and flanking sequence did not lead to ectopic en expression. The remaining PREs were apparently sufficient to recruit PcG proteins. However, we were surprised that loss of the DNA led to a loss-of-function phenotype, suggesting that the DNA must also play a positive role in the expression of en. Results of our current work suggest that the loss-of-function phenotype is attributable to the loss of a promoter-proximal tethering element (see below). In other experiments, we showed that PREs can either activate or repress transcription in a context-dependent manner. Further, our data suggest that PREs mediate looping between distant enhancers and the en promoter. Our experiments suggest activities of PREs not foreseen by others in the field.
The regulatory sequences for the en gene extend over a 70–kb region. Our laboratory used reporter constructs to find sequences important for expression in stripes, the nervous system, and head, among others. Discrete regulatory elements are located throughout the 70–kb region. We also found at least seven additional potential PREs located throughout the region. Others have shown that PcG protein complexes bring together DNA fragments in vitro, and it is possible that the complexes cause looping in vivo. We are interested in learning whether the additional PREs are involved in mediating interactions between distant enhancers and the en promoter.
The en gene exists in a gene complex with the nearby gene encoding invected (inv). The two genes are co-regulated and express proteins with largely redundant functions. By studying the regulation of en and inv, we are trying to understand how regulatory sequences up to 80 kb away regulate the activity of two promoters. Our current data suggest that PcG proteins bind to en DNA in all cells, even those that actively transcribe engrailed. We hypothesize that the chromatin modifications put down by the PcG proteins may be required for the activity of some en enhancers.
A genetic screen reveals the inhibitory effect of transcriptional activators on PRE activity.
We performed a genetic screen to identify new members of the Trithorax group and Polycomb group. The generation of transgenic Drosophila relies on the eye color gene white to detect transgenic flies. Eye color depends on the expression level of the white gene; more expression of white causes a darker eye color, whereas less expression causes a lighter eye color. PREs linked to white (a PRE–white transgene) cause less white expression, leading to a lighter eye color. We performed a genetic screen to identify mutations that darken the eye color of transgenic flies with a PRE–white transgene. We reasoned that mutation of a PcG gene that encodes a repressor might lead to darkening of the eye color. Increasing the activity of an activator protein (a potential Trithorax-group gene) might also darken eye color through competition with the PcG repressors. We screened over 60,000 flies and obtained nine mutants. We have now characterized two of the mutants and describe one below.
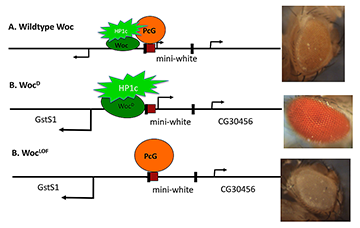
We obtained a dominant mutation in the transcriptional activator Woc (2). As shown by others, Woc stimulates transcription through an interaction with the protein HP1c. Our WocD mutant contains a single amino acid change, which may increase its activity. Our results suggest that increasing the activity of Woc reduces the ability of PcG repressors to act (Figure 2). The data point to the interplay between repressors and activators in setting the correct expression levels of genes.
Figure 2. Amount of activator influences Polycomb-group repression.
A model depicting the effect of competition between the activator Woc-HP1c and the repressor PcG proteins on the expression of the mini-white gene. The mini-white gene is encoded on a transgene that has a PRE (red box) upstream and is flanked by P-element ends (black boxes). The two genes that flank the mini-white transgene, GstSt and CG30465, are also shown. In wild type, Woc-HP1c and PcG co-exist to give a low level of mini-white transcription and a yellow eye color. In the wocD mutant, more HP1c is recruited than in wild type, leading to a loss of PcG protein repression, a higher level of mini-white transcription, and red eye color. In a woc loss-of-function (LOF) mutant, no HP1c is recruited, and PcG proteins silence the transcription of mini-white, leading to white eye color.
Increasing cohesin binding stability counteracts PcG silencing in Drosophila.
Cohesin is made up of the proteins Smc1, Smc3, Rad21, and Stromalin (SA) and is important for sister chromatid cohesion and proper chromosome segregation during mitosis. In addition, cohesin and cohesin-associated proteins play an important role in regulating gene expression. In a recent study, others found that the cohesin subunits Smc1, Smc3, and Rad21 co-purify with the PcG protein Polycomb (Strübbe et al., Proc Natl Acad Sci USA 2011;108:5572), suggesting that these protein complexes may physically interact at some loci. Wapl protein regulates binding of the cohesin complex to chromosomes during interphase and helps remove cohesin from chromosomes at mitosis. We isolated a dominant mutation in wapl (waplAG) in a screen for mutations that counteract silencing mediated by an engrailed PRE (3). waplAG hemizygotes die as pharate adults and have an extra sex combs phenotype characteristic of males with mutations in PcG genes (Figure 3). The wapl gene encodes two proteins, a long form and a short form. waplAG introduces a stop codon at amino acid 271 of the long form and produces a truncated protein. The expression of a transgene encoding the truncated Wapl-AG protein causes an extra-sex-comb phenotype similar to that seen in the waplAG mutant. Mutations in the cohesin-associated genes Nipped-B and pds5 suppress and enhance waplAG phenotypes, respectively. A Pds5–Wapl complex (releasin) removes cohesin from DNA, while Nipped-B loads cohesin, suggesting that Wapl–AG might exert its effects through changes in cohesin binding. Consistent with this model, Wapl-AG was found to increase the stability of cohesin binding to polytene chromosomes. Our data suggest that increasing cohesin stability interferes with PcG silencing at genes that are co-regulated by cohesin and PcG proteins.
Enhancer-promoter communication
Click image to enlarge.
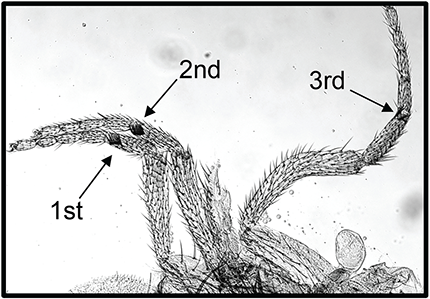
Figure 3. Wapl-AG causes extra sex comb teeth, the defining feature of PcG mutants.
This waplAG pharate adult male has sex comb teeth on all three legs (arrows). The second leg has 8 sex comb teeth, and the third leg has 2 sex comb teeth.
Enhancers are often located tens or even hundreds of kb away from their promoter, sometimes even closer to promoters of genes other than the one they activate. We showed that en enhancers can act over large distances, even skipping over other transcription units, choosing the en promoter over those of neighboring genes. Such specificity is achieved in at least three ways. First, early-acting en stripe enhancers exhibit promoter specificity. Second, a proximal promoter-tethering element is required for the action of the imaginal disk enhancer(s). Our data point to two partially redundant promoter-tethering elements. Third, the long-distance action of en enhancers requires a combination of the en promoter and sequences within or closely linked to the promoter-proximal PREs. The data show that several mechanisms ensure proper enhancer-promoter specificity at the Drosophila en locus, providing one of the first detailed views of how promoter-enhancer specificity is achieved.
Publications
- Brown JL, Kassis JA. Spps, a Drosophila Sp1/KLF family member, binds to PREs and is required for PRE activity late in development. Development 2010;137:2597-2602.
- Noyes A, Stefaniuk C, Cheng Y, Kennison JA, Kassis JA. Modulation of the activity of a Polycomb-group response element in Drosophila by a mutatoin in the transcriptional activator Woc. G3 (Bethesda) 2011;1:471-478.
- Cunningham MD, Gause M, Cheng Y, Noyes A, Dorsett D, Kennison JA, Kassis JA. Wapl antagonizes cohesin bindng and promotes Polycomb group silencing in Drosophila. Development 2012;139:4172-4179.
- Cheng Y, Kwon DY, Arai AL, Mucci D, Kassis JA. P-Element homing is facilitated by engrailed Polycomb-group Response Elements in Drosophila melanogaster. PLoS One 2012;7:e30437.
- Kassis JA. Transvection in 2012: Site-specific transgenes reveal a plethora of trans-regulatory effects. Genetics 2012;191:1037-1039.
Collaborators
- Dale Dorsett, PhD, Saint Louis University, St. Louis, MO
- Maria Gause, PhD, Saint Louis University, St. Louis, MO
- James A. Kennison, PhD, Program in Genomics of Differentiation, NICHD, Bethesda, MD
Contact
For more information, email jk14p@nih.gov.