You are here: Home > Section on Intracellular Protein Trafficking
Molecular Genetics of Neural Stem Cells

- Sohyun Ahn, PhD, Head, Unit on Developmental Neurogenetics
- Cheol Lee, PhD, Visiting Fellow
- Hui Wang, PhD, Postdoctoral Intramural Research Training Award Fellow
- Sherry Ralls, BA, Technician-Biologist
- Anna Kane, BS, Predoctoral Intramural Research Training Award Fellow
- Garrett Cheung, BS, Postbaccalaureate Intramural Research Training Award Fellow
Neural stem cells (NSCs) in the brain reside in specialized microenvironments called niches and produce new neurons throughout the life of an animal. Our overall goal is to understand how the NSCs are established during development and maintained after birth. Using the mouse as a model system, we study the interaction between NSCs and other cells within the neurogenic niche. Combining the power of genetics with other in vivo manipulation techniques, we are currently investigating the nature of regulatory signals presented by other cells in the niche, signals that dictate the behavior of NSCs. Insights gained from these studies should allow the development of cell type–specific strategies for the recruitment of endogenous NSCs during regeneration or for treatment of disease.
The role of Shh signaling and Gli3 in neurogenesis
Click image to enlarge.
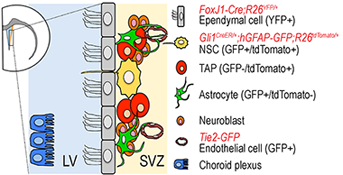
Figure 1. The neurogenic niche in adult subventricular zone of lateral ventricle in mouse forebrain
Various cell types constitute the neurogenic niche of the SVZ. We use a combination of transgenic and gene-targeted reporter mouse lines to isolate each cell type for gene expression profiling.
Sonic hedgehog (Shh) promotes the proliferation of NSCs in the adult brain. However, Shh signaling does not act on NSCs until late gestational stages, suggesting that proliferation of embryonic NSCs (also known as radial glia cells) and postnatal NSCs are regulated differently. The postnatal neurogenic niche contains various cell types, such as ependymal cells, that are also derived from embryonic NSCs around birth. However, it is still unclear how the distinct niche cell types are specified. To better understand the molecular–regulatory mechanism of Shh signaling, we focus on the Gli family of transcription factors (Gli1, Gli2, Gli3), which are activated or modified in response to Shh activity. Gli3 in particular is processed into a repressor form (Gli3R) in the absence of Hedgehog (Hh) signal and acts as the major negative transducer of Hh pathway. We previously showed that Gli3 plays a critical role in cortical neurogenesis, especially in the regulation of neural progenitors' proliferation and cell fate specification (1).
Click image to enlarge.
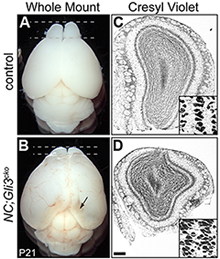
Figure 2. Reduced olfactory bulb in the absence of Gli3
21-day old mouse that lacks Gli3 function in developing nervous system (NC;Gli3cko) has smaller olfactory bulbs (B,D) than that of the control littermate (A, C).
During development, Shh signaling is also absent from embryonic neural stem cells. We demonstrated that embryonic radial glia cells require Gli3R to correctly specify postnatal ependymal cells and NSCs. Moreover, postnatal ependymal cells require Gli3R to maintain their cell identity, whereas the factor is not required in postnatal NSCs. In addition to the niche structure, proper neurogenesis depends on the continuous presence of Gli3R in ependymal cells, but not in NSCs, suggesting a cell non-autonomous (not restricted to the affected cell) requirement of Gli3R in the subventricular zone (SVZ), as evidenced by the smaller olfactory bulbs in Gli3 conditional mutants (Figure 2). We also found in vivo evidence that, in ependymal cells, Gli3R maintains a normal level of Numb, a negative regulator of the well characterized juxtacrine Notch signal pathway. Loss of Numb in postnatal ependymal cells led to compromised neurogenic activity of neighboring NSCs, thus providing the basis for the cell non-autonomous role of Gli3 in SVZ neurogenesis. Our findings highlight the importance of the restricted expression of negative regulators of the Shh and Notch pathways (Gli3R and Numb, respectively) in the SVZ cell types. In addition, we identified several signaling pathways and their components that are altered in the absence of Gli3, providing additional insights into the mechanisms by which NSCs and their environment interact to promote proper neurogenesis.
Gene expression profiling of adult neural stem cells and their niche
To expand our knowledge of the molecular repertoire that controls NSC behavior in the SVZ, we carried out systematic profiling of signaling molecules present in distinct niche cell types. We focused on identifying autocrine/paracrine signaling molecules as well as cell-cell contact–mediated signaling molecules. We took advantage of the Genetic Inducible Fate Mapping system and transgenic mice and specifically marked and isolated NSCs and SVZ niche cells, including ependymal cells, transit amplifying progenitors (TAPs), astrocytes, and vascular endothelial cells. We then validated the identity and purity of sorted cells and confirmed that purified NSCs were able to self-renew and produce multiple neural cell types in vitro. Using the Signal Sequence Trap method, we obtained the Secretory Molecule Expression Profile (SMEP) of each cell type and of choroid plexus tissue and identified a total of 151 genes encoding secretory or membrane proteins. In addition, we obtained the NSC SMEP of potential molecules involved in secretory signals, using cDNA microarray technology. We identified and tested, in NSCs in vitro, the physiological function of novel niche-related genes, including Enpp2, Ttr, Sparcl1, and CPE. Taken together, our results provide the first systematic profiling of signaling molecules present in distinct or overlapping cell types in the SVZ niche cells (Lee et al., under revision). Now that we have identified novel components related to the niche, we are pursuing the physiological in vivo function of these molecules in regulating NSC behaviors.
The genetic lineage of midbrain dopaminergic neurons
Click image to enlarge.
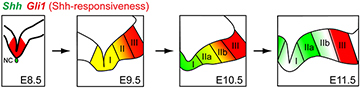
Figure 3. Dynamic nature of Sonic hedgehog (Shh) signaling in ventral midbrain
Shh expression and its responsiveness as read-out by Gli1 expression are dynamic during ventral midbrain development in the mouse. Only domains I and II produce midbrain dopaminergic neurons, not domain III.
The ventral midbrain (vMb) is organized into distinct anatomical domains and contains cohorts of functionally distinct subtypes of midbrain dopamine (mDA) neurons. The diverse molecular identities, spatial and anatomical distribution, and neural circuitry of mDA neurons give rise to the functional complexity of the neurons. We are interested in how the diverse mDA neurons are specified during embryonic development, with the focus on Shh signaling and using the mouse as a model system. We hypothesized that genetic history and timing of gene expression within mDA neuron progenitors regulate mDA neuron subtype identity. We tested this hypothesis using Genetic Inducible Fate Mapping to differentially mark the Shh and Gli1 lineages at various embryonic stages, and we demonstrated how timing and gene expression result in progenitor contribution to distinct mDA neuron domains. Importantly, we discovered that dynamic changes in Shh and Gli1 expression in the vMb primordia delineated Shh's and Gli1's spatial contribution to vMb. First, both lineages contributed to the medial domain. Subsequently, the Gli1 lineage exclusively contributed to the lateral vMb while the Shh lineage expanded more broadly across the vMb. Our findings show that the early Shh and Gli1 lineages specify mDA neurons of the substantia nigra pars compacta (SNc) while the late Shh and Gli1 lineages maintain their progenitor state longer in the posterior vMb to extend the production of mDA neurons in the ventral tegmental area (VTA). Taken together, our studies suggest that it is the timing of gene expression along with the genetic lineage (Shh or Gli1) within the neural progenitors that segregates mDA neurons into distinct mDA neuron subtypes (2).
Interestingly, the overall lineage contribution of both Shh and Gli1 lineages heavily overlaps among mDA neurons. A comparative expression analysis across the mesencephalic development showed that, at any given developmental stage, cells rarely co-express Shh and Gli1. Using ShhCre:GFP mice to delete the Smoothened receptor in the Shh pathway, we demonstrated that the loss of Shh signaling in Shh-expressing cells results in a transient rise in proliferation and subsequent depletion of mDA neuron progenitors in the posterior ventral mesencephalon (vMes). We found that this change in the proliferative behavior of vMes progenitors is caused by facilitated cell cycle exit. Moreover, the changes in duration of Shh signaling in vMEs progenitors altered the timing of the contribution to the VTA and SNc mDA neurons. Our investigation into the relationship between Shh–secreting and –responding cells reveals an intricate regulation of induction and cessation of Shh signaling that influences distribution of mDA neurons in the VTA and SNc (Hayes et al., under revision).
Additional Funding
- DOD Grant (2009-2011)
Publications
- Wang H, Ge G, Uchida Y, Luu B, Ahn S. Gli3 is required for maintenance and fate specification of cortical progenitors. J Neurosci 2011;31:6440-6448.
- Hayes L, Zhang Z, Albert P, Zervas M, Ahn S. The timing of Sonic hedgehog and Gli1 expression segregates midbrain dopamine neurons. J Comp Neurol 2011;519:3001-3018.
Collaborators
- Regina Armstrong, PhD, Uniformed Services University of the Health Service, Bethesda, MD
- Hwai-Jong Cheng, MD, PhD, University of California, Davis, CA
- Michael J. Holtzman, MD, Washington University, St. Louis, MO
- Y.P. Loh, PhD, Program in Developmental Neuroscience, NICHD, Bethesda, MD
- Yosuke Mukoyama, PhD, Genetics and Developmental Biology Center, NHLBI, Bethesda, MD
- Mark Zervas, PhD, Brown University, Providence, RI

