You are here: Home > Unit on Mammalian Epigenome Reprogramming
Epigenome Reprogramming During Mammalian Development

- Todd S. Macfarlan, PhD, Head, Unit on Mammalian Epigenome Reprogramming
- Carson Miller, PhD, Postdoctoral Fellow
- Elaine Bautista, BS, Postdoctoral Fellow
The overall goal of the laboratory is to understand how the epigenome is regulated in mammals during embryo development and to apply this knowledge to engineer tissues and build better models of disease. We explore how the genetic material is reprogrammed after fertilization to generate embryonic cells capable of generating all of the cell types that make up and support the embryo, and how these pluripotent cells are specified to particular neural fates. We utilize mouse genetics, developmental biology, biochemistry, cell biology, multi-dimensional imaging, stem cell–based in vitro modeling, and next-generation sequencing approaches to explore the role of histone-modifying enzymes and transcription factors during the reprogramming process.
Exploring the function of histone/DNA–modifying enzymes during development
A large number of enzymes that post-translationally modify histones/DNA have been identified and biochemically characterized. Many of these enzymes have also been shown to be essential for embryo development, mutated in cancers and neurological disease, or important for transgenerational epigenetic inheritance in model organisms. We are using stem cell–based models of development along with mouse genetics to remove enzymes that post-translationally modify histones and DNA in a tissue-specific manner. In particular, we are focusing on the period of preimplantation development, when massive reprogramming of the epigenome takes place, and during formation of the central nervous system, when cells are becoming post-mitotic as they differentiate into neurons and are thus incapable of replication-dependent changes in the epigenome. With these studies, we hope to gain a further understanding of the importance of epigenetic information stored in chromatin.
Interplay of transcription factors and chromatin-modifying enzymes
Transcription factors have long been known to be the master regulators of cell fate, binding to specific sequences of DNA and activating or silencing genes that allow specific cell types to carry out specialized function. More recently, it has been demonstrated that transcription factors can convert somatic cells into induced pluripotent cells (iPS cells) or from one somatic type to another, although this process is relatively inefficient. One likely reason that this process is inefficient is that differentiated cell types carry "restrictive" epigenomes that are less accessible to transcription factors than the "permissive" epigenomes of embryonic stem cells, which are easily reprogrammed to somatic cells. We are thus exploring the possibility that chromatin-modifying enzymes gate the activity of transcription factors during these reprogramming processes (both natural and artificial reprogramming), and we are testing how chromatin-modifying enzymes work in conjunction with transcription factors to regulate cell fate.
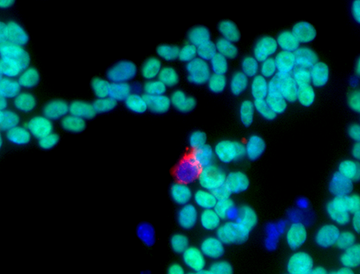
Figure 1. The murine endogenous retrovirus with tRNA leucine primer (MuERV-L) is expressed in a small population of cells within iPS cultures.
MuERV-L is transiently expressed in rare cells within pluripotent cells (labelled with antibodies that recognize the viral gag protein in red). The rare cells have a unique gene expression signature and functional characteristics in that they can contribute to both embryonic and extra-embryonic tissues in mouse chimeras. Because many of the genes that are also activated in this population use MuERV-L long-terminal repeat promoters, and similar viruses are found throughout placental mammals, we speculated that the virus has become an important regulator of cell fate within eutheria.
Exploring the regulation and function of endogenous retroviruses
Endogenous retroviruses (ERVs) are the remnants of ancient retroviral infections that have become a permanent part of the host genome. The sequences make up nearly 10% of mammalian genomes. It has long been debated whether ERVs and other transposable elements are nothing more than parasitic elements that have remained in genomes due to their ability to independently replicate (via retrotransposition) or whether they provide some selective advantage to their hosts. One way in which ERVs can influence their hosts is via regulation of host gene expression. ERVs and other transposons are targeted for silencing by distinct repressive chromatin–generating machinery that can spread to neighboring genes or can serve as alternate promoters. We have been studying a particular class of ERVs found in all placental mammals, called ERVLs (MERVLs in the mouse), that very briefly evade silencing in preimplantation development during zygote genome activation. Moreover, the ERVL elements appear to serve as primary or alternate promoters for many cellular genes, which led us to speculate that the elements may be essential for embryo development. We are interested in two important questions: (i) how ERVL elements are regulated; and (ii) whether ERVL elements are critical for cell fate decisions in embryo development.
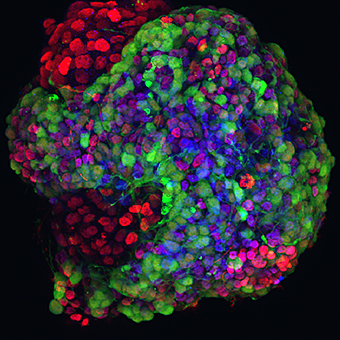
Figure 2. Embryonic stem cell–derived motor neurons
Motor neurons (labeled with an HB9::GFP reporter) were generated by differentiation of embryonic stem cells in vitro.
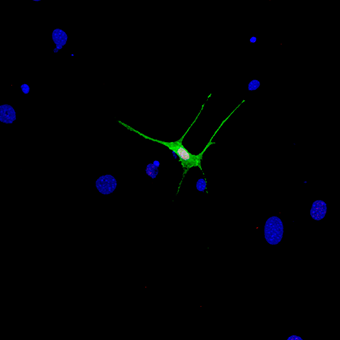
Figure 3. Transcription factor–mediated reprogramming of a fibroblast to a motor neuron
Primary fibroblasts were infected with lentiviruses expressing transcription factors specific for the motor neuron lineage. Fibroblasts undergoing reprogramming to motor-neuron–like cells are labeled with a green fluorescent protein.
Publications
- Macfarlan TS, Gifford WD, Driscoll S, Rowe HM, Bonanomi D, Lettieri K, Firth A, Singer O, Trono D, Pfaff SL. ES cell potency fluctuates with endogenous retrovirus activity. Nature 2012;487:57-63.
Collaborators
- Kai Ge, PhD, Laboratory of Endocrinology and Receptor Biology, NIDDK, Bethesda, MD
- Samuel Pfaff, PhD, The Salk Institute and the Howard Hughes Medical Institute, La Jolla, CA
- Anjana Rao, PhD, La Jolla Institute for Allergy and Immunology, La Jolla, CA
- Bing Ren, PhD, University of California San Diego, San Diego, CA
- Arthur Skoultchi, PhD, Albert Einstein College of Medicine, New York, NY
- Azim Surani, PhD, Gurdon Institute, University of Cambridge, Cambridge, UK
- Didier Trono, MD, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Contact
For more information, email lovep@mail.nih.gov.