You are here: Home > Section on Vertebrate Development
Control of Ectodermal Development in Vertebrate Embryos

- Thomas D. Sargent, PhD, Head, Section on Vertebrate Development *
- Hiu Wan Law, Visiting Fellow
- Yoko Ogawa, PhD, Visiting Fellow
- Valerie Virta, PhD, Postdoctoral Intramural Research Training Award Fellow
* Dr. Sargent retired in 2016.
The primary interest of this group is the control of cranial neural crest (CNC) development. We started the project by identifying the transcription factor TFAP2 as a critical component of NC induction in the frog Xenopus, then went on to study regulatory targets of TFAP2, including a novel protein we named Inka. Inka acts as a regulator of cytoskeletal dynamics, partly by interacting with a protein kinase, Pak4. The kinase is known to mediate cell-cell signals that influence cell shape, adhesion, and migration, both in development and in cancer. The focus of the Inka project has shifted to Pak4 function in zebrafish. Knockdown experiments with antisense morpholinos revealed that Pak4 functions as an essential maternal effect gene in zebrafish. In contrast, zygotic expression of the zebrafish pak4 gene appears to be dispensable. We are currently using genome-level approaches to continue this investigation.
We also initiated two new CNC projects, also being carried out in zebrafish: one is to use the mutation dlx3TDO in the homeobox gene dlx3. The mutation is the basis for a human genetic disorder affecting bones, teeth, and hair, and converts the transcription factor into a dominant-negative inhibitor of wild-type Dlx3. We are probing the function of Dlx3 in zebrafish using this molecule as a tool. The other project is to manipulate cell-cell signaling in migrating CNC cells in the living embryo by experimentally regulated expression of transgenes encoding engineered signaling molecules.
Pak4 function in zebrafish
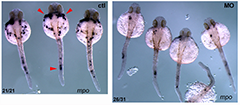
Click image to enlarge.
Figure 1. Pak4 knockdown inhibits neutrophil development.
Knockdown of zebrafish Pak4 by morpholino oligonucleotides drastically reduces the formation of neutrophils. Visualized by in situ hybridization with a probe for myeloperoxidase (mpo; red arrowheads), a neutrophil-specific gene.
The human gene encoding PAK4 (p21-activated kinase 4) belongs to a family of six genes. PAKs bind to small GTPase effector molecules—the Rho-class proteins Cdc42 and Rac—and transduce signals to the cytoskeleton and nucleus, affecting cell shape and gene expression, respectively. Overexpression of Pak4 is tumorigenic in mouse cells, and the abundance of PAK4 in many tumors suggests that it mediates a common step in cancer progression. In the mouse, disruption of the Pak4 gene yields a complex embryonic lethal phenotype affecting vasculature, heart, and neuronal and other tissues. Our work in zebrafish showed that pak4 in this species is most strongly expressed in the egg, i.e., maternally, and that maternal expression is essential whereas the zygotic pak4 appears to dispensable, at least through the first week or so of life. Loss of maternal pak4 in zebrafish results in a complex lethal phenotype, including abnormal axial muscle development and defects in hematopoiesis—most strikingly, the suppression of neutrophils, cells involved in inflammatory response to infection or injury (Figure 1). Other tissues, including the craniofacial cartilage, were also affected in the knockdown embryos. Given that PAK4 is an established regulator of cytoskeletal dynamics, we tested pharmacological cytoskeleton filament disuptors for effects on the genes we found to be altered by pak4 knockdown. Drugs leading to actin filament depolymerization seem to duplicate, at least in part, the effects of losing maternal pak4. The findings reveal a novel connection between the cytoskeleton and the regulation of specific developmental transcription programs. We are continuing to investigate this question and are also generating zebrafish with mutations in the pak4 gene using TALENs, which are target-specific nucleases capable of disrupting single copy genes in zebrafish (and other) embryos.
Dlx gene function in cranial neural crest development
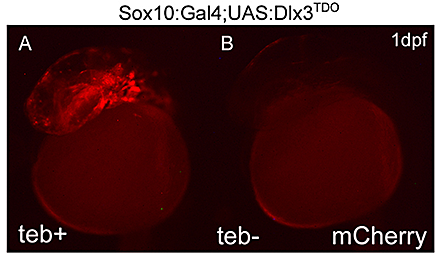
Click image to enlarge.
Figure 2. Expression of DLX3TDO in the early neural crest
A: 24-hour zebrafish embryos transgenic for both the sox10:Gal4EcR driver and UAS:DLX3TDO target genes, treated with tebufenozide beginning at shield stage. B: An untreated double transgenic embryo demonstrating the inducible system. The red fluorescence is from mCherry, co-expressed with DLX3 using viral 2A peptide linkage.
The distal-less class of homeobox genes have been linked for some time with the regulation of cranio-facial morphology. Of this group of six genes (Dlx1–6 in mammals), Dlx1, Dlx2, Dlx5, and Dlx6 have been most extensively investigated in this context. We showed in Xenopus that dlx3 expression is excluded from CNC at early stages, that this could be attributed to differential sensitivity to local BMP signal strength, and that ectopic expression of dlx3 (but not other dlx genes) in the early CNC obliterated this differentiation pathway. Knockdown of dlx3b in zebrafish predominantly leads to defects in the otic (ear) placode. However, combined knockdown of dlx3b and other dlx genes results in multiple jaw defects, indicating redundant functions. In humans, the dominant autosomal genetic disorder tricho-dento-osseous syndrome (TDO) affects some aspects of CNC development, and it is thought that this occurs by a dominant-negative effect on the wild-type DLX3 allele, or possibly on other members of this superfamily (DlX and related MSX factors can form heterodimers in vivo). In the zebrafish, we are using the naturally occurring mutant dlx3 to inhibit the wild-type Dlx3 protein and potentially other co-expressed dlx genes in the CNC. It is important to maintain spatial and temporal control of expression for this approach. To achieve this, we are using a hormone (ecdysone)-inducible fusion of a modified Gal4 transcriptional activator, driven in transgenic zebrafish embryos with the promoter of the sox10 gene, which is neural crest–specific at early stages of development. This is used in conjunction with zebrafish carrying a transgene with an upstream activating sequence (UAS) linked to the human DLX3TDO gene (Figure 2). Preliminary results suggest that zebrafish expressing DLX3TDO exhibit malformations in certain bones and cartilages in the head. We will continue these experiments using different time windows of DLX3TDO expression to identify the critical period for craniofacial morphological specification. We are also developing a similar transgenic approach using a synthetic dlx3b gene construct, which includes the Drosophila engrailed repressor domain and which we predict will have much stronger effects on CNC development. Our findings will be interpreted in the light of equivalent gene replacement experiments that our collaborator Maria Morasso is carrying out in the mouse.
Cell-cell signaling in cranial neural crest development
Bmp4 has been identified as a crucial signal for determining mouth shape in vertebrates. To extend our earlier studies, we will use the binary sox10:GalrEcR'-UAS system outlined in the previous section to target expression of a truncated dominant negative type I bmp receptor to the NC lineage. For transgenesis, in addition to our lab standard EK line, we are using a background line that expresses a membrane-bound RFP under sox10 control, to facilitate imaging of craniofacial elements at the cellular level. Production of the target transgenic lines is currently under way. When suitable lines have been generated and characterized, we will cross them with the sox10:Gal4EcR driver line and induce expression by tebufenozide. We will collect data in three dimensions using confocal microscopy so that we can make comparisons between control and experimentally manipulated embryos. The ability to control the timing and location of expression of the BMP signaling inhibitor, along with advanced three-dimensional live imaging, is expected to provide a long-term strategy for evaluating signaling in migratory and postmigratory cranial NC development. Ideally we should be able to induce predictable structural changes in craniofacial morphology and correlate them with changes in gene expression.
Publications
- Law HW, Sargent TD. Maternal expression of Pak4 is required for primitive myelopoiesis in zebrafish. Mech Dev 2012;in press.
Collaborators
- Maria Morasso, PhD, Laboratory of Skin Biology, NIAMS, Bethesda, MD
Contact
For more information, email sargentt@mail.nih.gov or visit sargentlab.nichd.nih.gov.

