You are here: Home > Section on Chromatin and Gene Expression
Chromatin Remodeling and Gene Activation

- David J. Clark, PhD, Head, Section on Chromatin and Gene Expression
- Hope A. Cole, PhD, Postdoctoral Fellow
- Peter Eriksson, PhD, Research Fellow
- Dwaipayan Ganguli, PhD, Visiting Fellow
- Nagarajavel Vivekananthan, PhD, Visiting Fellow
Gene activation involves the regulated recruitment of factors to a promoter in response to appropriate signals, ultimately resulting in the formation of an initiation complex by RNA polymerase II and subsequent transcription. These events must occur in the presence of nucleosomes, which are compact structures capable of blocking transcription at every step. To circumvent the chromatin block, eukaryotic cells possess chromatin-remodeling and nucleosome-modifying complexes. The former (e.g., the SWI/SNF complex) use ATP to drive conformational changes in nucleosomes and to slide nucleosomes along DNA. The latter contain enzymatic activities (e.g., histone acetylases) that modify the histones post-translationally to mark them for recognition by other complexes. Our previous work using native plasmid chromatin from the yeast Saccharomyces cerevisiae revealed that activation correlates with large-scale movements of nucleosomes and conformational changes within nucleosomes over entire genes. Using paired-end sequencing, we have now confirmed and extended these observations by determining nucleosome positions genome-wide. In addition, we discovered that, unlike canonical nucleosomes, the centromeric nucleosome, which contains a specialized histone (Cse4/CenH3), is perfectly positioned. Our current work focuses on the roles of chromatin-remodeling machines in nucleosome positioning and gene activation.
Transcriptional activation, nucleosome mobilization, and nucleosome position clusters
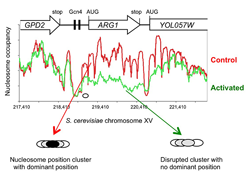
Click image to enlarge.
Figure 1. Activation of the yeast ARG1 gene with 3-aminotriazole results in dramatic loss of nucleosome occupancy and disruption of nucleosome positioning over the coding region, which extends into the downstream YOL057W gene.
An example of genome-wide data obtained using paired-end sequencing (2). Oval: nucleosome drawn to scale.
We chose budding yeast as a model organism because biochemical studies of chromatin structure could be combined with molecular genetics. Current models for the role of the SWI/SNF ATP–dependent chromatin remodeling complex in gene regulation focus on promoters, where the most obvious changes in chromatin structure occur. However, using our plasmid model system with HIS3, an SWI/SNF–regulated gene, we discovered that transcriptional activation creates a domain of remodeled chromatin structure that extends far beyond the promoter to include the entire gene. We addressed the effects of transcriptional activation on the chromatin structure of HIS3 by mapping the precise positions of nucleosomes in non-induced and transcriptionally activated chromatin. In the absence of the Gcn4 activator, the HIS3 gene is organized into a dominant nucleosomal array. In wild-type chromatin, the array is disrupted, and several alternative, overlapping nucleosomal arrays form. Disruption of the dominant array also requires the SWI/SNF remodeling machine, indicating that the SWI/SNF complex plays an important role in nucleosome mobilization. We suggest that the net effect of interplay among remodeling machines at HIS3 is to create a highly dynamic chromatin structure (1). Our work on HIS3 and our earlier work on CUP1 indicate that, at least for these two genes, the target of remodeling complexes is a domain rather than just the promoter. This important finding suggests that remodeling complexes act on chromatin domains. We speculate that remodeling entire genes might facilitate elongation through nucleosomes by RNA polymerase II.
Click image to enlarge.
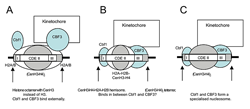
Figure 2. Models for the perfectly positioned centromeric nucleosome (3)
A. The CenH3 histone octamer model in which CenH3 replaces H3 in an otherwise canonical histone octamer. B. The hemisome model in which a CenH3-H4-H2A-H2B tetramer is bound tightly to sequence-specific factors Cbf1 and CBF3. C. A tetramer containing two molecules of CenH3 and H4 is bound tightly to Cbf1 and CBF3.
The advent of high-throughput sequencing has allowed us to analyze nucleosome positioning on a genome-wide scale, confirming and extending our previous observations. In collaboration with Bruce Howard, we are using paired-end sequencing, which increases the accuracy of position measurements and facilitates bioinformatics analysis. We showed that major disruptions occur in the chromatin structure of genes when they are activated for transcription, including a dramatic loss of canonical nucleosomes and a re-positioning of the remaining nucleosomes (2). An example is the ARG1 gene (Figure 1). We also discovered that, unlike canonical nucleosomes, which occupy one of several alternative overlapping positions, the specialized nucleosome at the centromere is essentially perfectly positioned (3). That is, it occupies exactly the same position with respect to the centromeric DNA sequence in all cells. We attribute the perfect positioning of the centromeric nucleosome to the presence of sequence-specific factors built into the nucleosome (Figure 2). Currently, we are exploring the roles of remodeling complexes capable of nucleosome mobilization in disruption of chromatin structure.
A nucleosomal barrier to transcription by RNA polymerase II in vitro and in vivo
Many eukaryotic genes are regulated at the level of transcript elongation. Nucleosomes are likely targets for such regulation. Previously, the Studitsky laboratory showed that nucleosomes formed on very strong positioning sequences (601 and 603) present a high, orientation-dependent barrier to transcription by RNA polymerase II in vitro. The existence of this polar barrier correlates with the interaction of a 16-bp polar barrier signal (PBS) with the promoter-distal histone H3-H4 dimer. In collaboration with the Studitsky and the Bartholomew laboratories, we found that the polar barrier is relieved by ISW2, an ATP-dependent chromatin remodeler that translocates the nucleosome over a short distance, such that the PBS no longer interacts with the distal H3-H4 dimer, although it remains within the nucleosome. In vivo, insertion of the 603 positioning sequence into the yeast CUP1 gene results in a modest reduction in transcription, but the reduction is orientation-independent, indicating that the polar barrier may be circumvented. Surprisingly, the 603 nucleosome is present at the expected position in only a small fraction of cells. Thus, the polar barrier is probably non-functional in vivo because the nucleosome is not appropriately positioned, presumably in response to nucleosome sliding activities. Interactions between polar barriers and chromatin remodelers might have significant regulatory potential (4).
Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast.
Click image to enlarge.
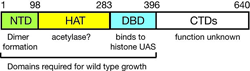
Figure 3. Domain structure of Spt10p
NTD = N-terminal domain; HAT = histone acetylase; DBD = DNA-binding domain; CTDs = C-terminal domains; UAS = upstream activating sequences
Previously, we showed that induction of the CUP1 gene results in acetylation of nucleosomes at the CUP1 promoter. The acetylation depends on SPT10, which encodes a putative histone acetylase (HAT) and has been implicated as a global regulator. Using expression microarrays, we confirmed that Spt10 has global effects on transcription. Surprisingly, the global effects of Spt10 are the indirect result of defects in chromatin structure, reflecting SPT's role as activator of the core histone genes. We demonstrated that Spt10 binds specifically to pairs of upstream activating sequences (UAS elements) in the core histone promoters (consensus: (G/A)TTCCN6TTCNC), consistent with a direct role in histone gene regulation. The DNA–binding domain of Spt10 contains an unusual zinc finger with homology to foamy retrovirus integrase, which we propose is a sequence-specific DNA–binding protein. Thus, Spt10 is a highly unusual trans-activator in which a HAT domain, normally recruited as a co-activator to promoters through an activation domain, is attached directly to a sequence-specific DNA–binding domain (Figure 3).
Click image to enlarge.
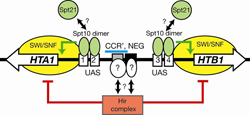
Figure 4. Regulation of the HTA1-HTB1 locus
An Spt10p dimer binds at each pair of UAS elements and activates transcription when a cell cycle–regulated signal is received (which could be Spt21p, or phosphorylation by a cell cycle–dependent kinase). Activation requires SWI/SNF and is repressed by the Hir complex; these effects require the NEG region (blue bar), which contains CCR′ and the NEG element (black box), both of which bind to unidentified sequence-specific factors.
We addressed the role of Spt10 in the cell cycle–dependent regulation of the histone genes. Such regulation is necessary to provide histones for nucleosome assembly during DNA replication. Histones H2A and H2B are expressed from divergent promoters at the HTA1-HTB1 and HTA2-HTB2 loci. We showed that Spt10 binds to two pairs of UAS elements in the HTA1-HTB1 promoter: UAS1/UAS2 drive HTA1 expression and UAS3/UAS4 drive HTB1. UAS3 and UAS4 also contain binding sites for the cell cycle regulator SBF (an Swi4-Swi6 heterodimer), which overlap the Spt10 binding sites. Spt10 and SBF binding to UAS3 and UAS4 is mutually exclusive in vitro. Both SBF and Spt10 are bound in cells arrested with alpha-factor, apparently awaiting a signal to activate transcription. Soon after removal of alpha-factor, SBF initiates a small, early peak of HTA1 and HTB1 transcription, followed by a much larger peak attributable to Spt10. Both activators dissociate from the HTA1-HTB1 promoter after expression has been activated. Thus, SBF and Spt10 cooperate to control the timing of HTA1-HTB1 expression (5).
Our current work aims to identify negative regulatory proteins that bind at the histone promoters, which might counteract activation by Spt10 (Figure 4) and to understand how the positive (UAS) and negative regulatory systems interact during the cell cycle.
Publications
- Clark DJ. Nucleosome positioning, nucleosome spacing and the nucleosome code. J Biomol Struc Dyn 2010;27:781-793.
- Cole HA, Howard BH, Clark DJ. Activation-induced disruption of nucleosome position clusters on the coding regions of Gcn4-dependent genes extends into neighbouring genes. Nucleic Acids Res 2011;[Epub ahead of print].
- Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc Natl Acad Sci USA 2011;108:12687-12692.
- Gaykalova DA, Nagarajavel V, Bondarenko VA, Bartholomew B, Clark DJ, Studitsky VM. A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation. Nucleic Acids Res 2011;39:3520-3528.
- Eriksson PR, Ganguli D, Clark DJ. Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast. Mol Cell Biol 2011;31:557-572.
Collaborators
- Vasily M. Studitsky, PhD, University of Medicine and Dentistry of New Jersey, Piscataway, NJ
- Bruce H. Howard, MD, Program on Genomics of Differentiation, NICHD, Bethesda, MD
- Blaine Bartholomew, PhD, Southern Illinois University, Carbondale, IL
Contact
For more information, email clarkda@mail.nih.gov or visit ucge.nichd.nih.gov.

