You are here: Home > Section on Molecular Genetics of Immunity
Gene Regulation in Innate Immunity

- Keiko Ozato, PhD, Head, Section on Molecular Genetics of Immunity
- Anup Dey, PhD, Biologist
- Tomohiko Kanno, MD, PhD, Staff Scientist
- Natarajan Ayithan, PhD, Visiting Fellow
- Mira Patel, PhD, Visiting Fellow
- Naoyuki Sarai, PhD, Visiting Fellow
- Monica Gupta, PhD, Visiting Fellow
- Songxiao Xu, PhD, Visiting Fellow
- Yuko Yoshida, PhD, Visiting Fellow
- Walter Huynh, BS, Postbaccalaureate Intramural Research Training Award Fellow
- Daniel Kim, BS, Postbaccalaureate Intramural Research Training Award Fellow
Macrophages (MФ) and dendritic cells (DC) respond to pathogen stimuli and produce cytokines including interferons (IFNs), IL-1, IL-6, and TNF-alpha to impart anti-viral and anti-microbial status to the host. Our goal is to study molecular pathways that direct the development and function of MФ and DC. Our long-term interest has been the role of a transcription factor IRF8 in innate immunity. IRF8 is expressed in MФ and DC at high levels and is required for production of both type I and type II IFNs. IRF8 and related transcription factors including IRF3 and IRF7 are post-translationally modified by ubiquitin and small ubiquitin-like molecules (SUMO). SUMO modification of these factors almost always results in transcriptional repression. SUMO–mediated repression likely represents an important mechanism to limit overproduction of inflammatory cytokines.
Click image to enlarge.
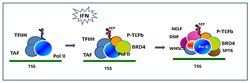
Figure 1. BRD4–dependent assembly of elongation factors
Interferon (IFN)–induced recruitment of elongation factors. IFN stimulation activates the JAK/STAT pathway and triggers histone acetylation and recruitment of BRD4 on the transcription start site (TSS) of IFNH–stimulated genes. BRD4 brings in P-TEFb that phosphorylate RNA polymerase II. The BRD4/P-TEFb recruitment in turn stimulates recruitment of additional elongation factors, NELF/DSIF and SPT6, to accomplish productive elongation.
Transcriptionally active genes are embedded in chromatin that is dynamically exchanged, whereas silenced genes are surrounded by more stable chromatin. The chromatin environment influences transcriptional processes and controls epigenetic regulation. We are interested in BRD4, a chromatin-binding protein associated with transcribed genes. We are also interested in histone H3.3, the variant histone that is selectively associated with actively expressed genes. BRD4 is a 200 kDa nuclear protein carrying two tandem bromodomains through which it binds to acetylated chromatin. BRD4 also interacts with the elongation factor P-TEFb and regulates transcription of many genes, including those induced by external stimuli. BRD4 is implicated in transcriptional memory across cell division because it stays on condensed chromosomes during mitosis and affects gene expression in the daughter cells. Despite close structural similarity with the standard H3.1 and H3.2, histone H3.3 has an extraordinary property, in that it is incorporated into nucleosomes and DNA only in actively transcribed genes. In contrast, H3.1, H3.2, and other standard core histones are incorporated into nucleosomes during DNA replication. The difference between H3.3 and H3.1/2 reflects distinct chromatin activities during replication and transcription. Although there is mounting recognition of the importance of transcription-coupled histone incorporation, the process and its physiological significance is still shrouded in mystery. Our goal is to elucidate the activity of BRD4 and histone H3.3 in the context of transcriptional activation and epigenetic memory.
Ubiquitin and SUMO modifications of nuclear proteins during innate immune responses
Because IRF8 and several other IRF members are modified by both ubiquitin and SUMO, we investigated whether these modifications occur in other proteins during innate immune responses. Macrophages were stimulated with IFNg and a toll-like receptor (TLR) ligand, CpG, which activates the TLR pathway and triggers transcription of many genes important for macrophage function. Some of these genes are responsible for resistance to pathogens, and other genes are involved in inflammatory responses in the host. To detect global ubiquitin or SUMO modifications of nuclear proteins in stimulated macrophages, we performed immunoblot analysis using antibody against ubiquitin, SUMO1, or SUMO2/3, testing nuclear fractions obtained from stimulated RAW264.7 macrophage cells. While there is only one ubiquitin protein, there are three SUMO molecules. SUMO1 is distinct from SUMO2 and SUMO3, which are more than 90% identical in amino acid sequence; currently, there is no antibody that distinguishes SUMO2 from SUMO3. We found a dramatic increase in ubiquitin-linked proteins following IFNg/CpG stimulation. IFNg treatment alone resulted in a significant increase in ubiquitin-linked proteins. However, additional stimulation by CpG further increased the amount of ubiquitin-bound proteins. Not only CpG, but other TLR ligands tested, including LPS and poly IC, also caused increased ubiquitination, provided that macrophages were prestimulated with IFNg. Similarly, IFNg/TLR stimulation caused a marked increase in proteins conjugated to SUMO1 or SUMO2/3. Our data demonstrate that ubiquitin and SUMO modifications are an integral part of innate immune responses, likely coupled with large changes in transcription and other nuclear activities, which helps mediate a rapid response to pathogen stimuli.
To begin cataloguing individual proteins modified by ubiquitin, we performed proteomic analysis of ubiquitin-bound proteins in stimulated macrophages. We stimulated RAW macrophages stably expressing Flag-tagged ubiquitin with IFNg/CpG and analyzed proteins co-precipitated with anti-Flag antibody by MSMS mass spectrometry. These proteins included factors in the ubiquitin pathways, those involved in DNA metabolism, chromatin and transcriptional regulation, and mRNA processing. The largest group of proteins found in our list were ribosomal proteins important for protein translation. Other proteins found were heat shock proteins and stress-response factors, suggesting a link between macrophage activation and the stress response. In conclusion, upon macrophage activation, a large number of nuclear proteins become associated with ubiquitin modification, presumably leading to a global shift in genome activity that is important for proper execution of innate immune responses.
The role of Brd4 in transcriptional memory during mitosis
Click image to enlarge.
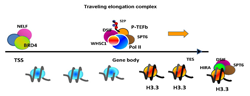
Figure 2. Molecular complexes that stay or travel during elongation
RNA Pol II, phosphorylated at serine 2 in the CTD, travels through the ISG gene body along with P-TEFb DSIF, SPT6, and WHSC1, which go beyond the transcription end site (TES). BRD4 and NELF, however, remain at the TSS. Elongation processes causes deposition of histone H3.3 preferentially in the gene body and at the TES.
Interaction of the chromatin-binding protein Brd4 with acetyl histones persists during mitosis, while its interaction with P-TEFb is interrupted during mitosis when most transcription factors, including P-TEFb, are released from chromatin. By detailed ChIP analysis of cells in mitosis, we found that Brd4 localized largely to the transcription start sites (TSS) of genes that are expressed immediately after the completion of mitosis. The genes have been classified as M-G1 genes and include Ran, Kif5b, Topo1, Tgif, Gnb1, Rad21, etc. Brd4 was largely absent from genes that are expressed later during the cell cycle, such as during late G1, S, and G2. Correlating with Brd4–binding patterns, levels of H3/H4 acetylation were higher in M-G1 genes than in those genes expressed later. While the bulk of RNA polymerase II was released from chromatin during mitosis, some was found at or near the TSS of most genes expressed in the cells. Furthermore, binding of Brd4 to the TSS dramatically increased at telophase, the last stage of mitosis, when post-mitotic transcription starts. The increased binding of Brd4 coincided with a sharp increase in histone H3/H4 acetylation, presumably catalyzed by reassembled histone acetylases. Correlating with this event, the real-time mobility of Brd4 temporarily declined during telophase, as measured by fluorescent recovery after photobleaching, indicating that the affinity of Brd4 for chromatin is elevated at telophase. We also noted that P-TEFb is recruited to the TSS of Brd4-bound M-G1 genes during telophase. This, in turn, coincides with the phosphorylation of the C-terminal domain of RNA polymerase II at serine 2, signifying active transcription of M-G1 genes. To address the importance of the binding of Brd4 to M-G1 genes, we knocked down Brd4 expression by shRNA and found that recruitment of P-TEFb and phosphorylation of RNA polymerase II was markedly reduced in Brd4–knockdown cells. Accordingly, Brd4–knockdown cells exhibited greatly impaired M-G1 gene expression. Our results indicate that Brd4 selectively marks M-G1 genes during mitosis and directs their transcription by mobilizing P-TEFb.
Transcriptional elongation and chromatin exchange in interferon-stimulated genes
BRD4 regulates genes involved in cell cycle progression from G0, G1, through S, thus affecting cell growth. Recently, it has been shown that BRD4 is required for induction of inflammatory genes in macrophages, suggesting a role in innate immunity. We investigated whether BRD4 plays a role in transcription of interferon-stimulated genes (ISGs). Type I IFN activates a large number of genes through the JAK/STAT pathway and establishes natural resistance against viruses and other pathogens. We found that IFN stimulation triggers recruitment of BRD4 to the TSS of a number of ISGs. Recruitment of BRD4 to ISGs coincided with increased histone acetylation at the TSS and induction of ISG mRNAs. Further, BRD4 recruitment coincided with the binding of the elongation factor P-TEFb, which is known to interact with BRD4. We also showed that the pausing complex of NELF and DSIF is recruited to the ISGs after IFN stimulation. IFN–induced NELF/DSIF recruitment was unexpected, given that the pausing complex has been reported to be present prior to stimulation and dissociate after activation, as studied for some inducible genes. Furthermore, another elongation factor, SPT6, known to function as a histone H3 chaperon, was recruited to ISGs after IFN stimulation. The results indicate that several elongation factors are induced to assemble on the ISGs. To delineate a hierarchical order of the assembly of the factors, we tested the effect of a small-molecule inhibitor that inhibits binding of acetylated histones to bromodomains. This inhibitor not only inhibited BRD4 recruitment, but also recruitment of P-TEFb, NELF/DSIF, and SPT6. In addition, the inhibitor markedly reduced ISG transcription. Similarly, shRNA-based BRD4 knockdown led to reduced recruitment of all the above factors. The data support the view that BRD4 recruitment is the primary event to initiate a cascade of factor binding that shapes overall ISG transcription. Additional knockdown studies showed that NELF and SPT6 have opposing activities in elongation, the former repressing ISG transcription while the latter promotes transcription. Together, our results indicate that BRD4 orchestrates ISG transcription by coordinating positive and negative elongation.
We also studied H3.3 incorporation into IFN–stimulated genes, using NIH3T3 cells expressing H3.3-fused to the yellow fluorescent protein (YFP). Following IFN stimulation, H3.3-YFP was rapidly incorporated into all four IFN–activated genes tested, with the highest enrichment seen in the distal end of the coding region. Surprisingly, H3.3 enrichment in the coding region continued for an extended period of time, long after transcription ceased. The promoter region, although constitutively enriched with H3.3-YFP, did not show an increase in H3.3-YFP deposition in response to IFN stimulation. Further, while H3.3-YFP deposition persisted in non-dividing cells for days after IFN stimulation, it rapidly declined in dividing cells. Lastly, we examined the role of H3.3 in IFN–stimulated transcription by an shRNA approach and found that IFN–stimulated transcription was significantly impaired in H3.3-knockdown cells. Our results indicate that H3.3 plays a role in IFN–mediated transcription and that its deposition leaves a prolonged post-transcriptional mark in these genes.
Additional Funding
- Trans NIH-FDA Biodefense Program, IATAP Program
Publications
- Kubota T, Matsuoka M, Xu S, Otsuki N, Takeda M, Kato A, Ozato K. Piasy inhibits virus-induced and interferon-stimulated transcription through distinct mechanisms. J Biol Chem 2011;286:8165-8176.
- Alsarraj J, Walker RC, Webster JD, Geiger TR, Crawford NP, Simpson RM, Ozato K, Hunter KW. Deletion of the proline-rich region of the murine metastasis susceptibility gene Brd4 promotes epithelial-to-mesenchymal transition- and stem cell-like conversion. Cancer Res 2011;71:3121-3131.
- Kim JY, Anderson ED, Huynh W, Dey A, Ozato K. Proteomic survey of ubiquitin-linked nuclear proteins in interferon-stimulated macrophages. J Interferon Cytokine Res 2011;31:619-628.
- Ouyang X, Zhang R, Yang J, Li Q, Qin L, Zhu C, Liu J, Ning H, Shin MS, Gupta M, Qi CF, He JC, Lira SA, Morse HC 3rd, Ozato K, Mayer L, Xiong H. Transcription factor IRF8 directs a silencing programme for TH17 cell differentiation. Nat Commun 2011;2:314.
- Liang Q, Deng H, Li X, Wu X, Tang Q, Chang T-H, Peng H, Rauscher FJ 3rd , Ozato K, Zhu F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier e3 ligase and negative regulator of IFN regulatory factor 7. J Immunol 2011;187:4754-4763.
Collaborators
- Toru Kubota, PhD, Japan National Institute of Infectious Diseases, Tokyo, Japan
- Ben-Zion Levi, PhD, Technion, Israel Institute of Technology, Haifa, Israel
- Herbert Morse II, MD, Laboratory of Immunopathology, NIAID, Rockville, MD
- Tomohiko Tamura, MD, PhD, Tokyo University, Tokyo, Japan
Contact
For more information, email ozatok@mail.nih.gov or visit ozatolab.nichd.nih.gov.

