You are here: Home > Section on Human Genetics
The Chromatin-Based Epigenome in Development and Aging

- Bruce H. Howard, MD, Head, Section on Human Genetics
- Valya Russanova, PhD, Staff Scientist
- Paraskevi Salpea, BS, Predoctoral Fellow
- Megan Cermak, BS, Postbaccalaureate Fellow
The normal human lifespan is marked by a complex series of developmental transitions, with relative stability during adulthood and, ultimately, a gradual decline in viability. Clock-like mechanisms presumably underlie the developmental events that occur through childhood and adolescence. Further, instabilities in such mechanisms are likely to be an integral part of the aging process and to contribute to many common degenerative diseases of later life. A breakthrough in the area of developmental transitions would have remarkable implications and underscores today's widespread interest in the rapidly evolving field of epigenomics as the key to further progress.
Whole-genome comparisons of epigenome structure
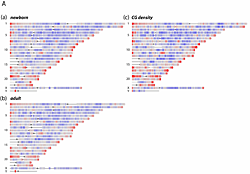
Click image to enlarge.
Figure 1. DNA methylation levels on a whole-genome scale
Sequence reads from precipitated MeDIP DNA were analyzed with a 2 Mb genomic window (1 Mb steps) and aligned to single-copy (RepeatMasker excluded) regions of the human genome, build 36.1. Red color represents <=2-fold over-representation, and blue <= 2-fold under-representation for MeDIP signals compared to input control DNA. (c) CG dinucleotide densities in single-copy regions relative to genome average.
The past year has seen the scaling up of high-throughput genomics approaches in studies that use human tissues from several clinical sources. We obtain peripheral blood monocytes from newborns (cord blood) through a collaboration with the Perinatology Branch, NICHD, while monocytes from adults are available through the NIH Department of Transfusion Medicine. The cells are analyzed either directly or after differentiation in vitro into antigen-presenting dendritic cells. We procure human skin fibroblasts from newborns and adults, as needed, under a protocol approved by the NICHD Institutional Review Board. We induce the latter cells to enter a quiescent state by serum deprivation for six or more days and then examine them with respect to gene expression and chromatin structure upon serum stimulation.
With both monocyte- and fibroblast-based experimental systems, we previously studied developmental and age-related changes in gene regulation. Our current work employs next-generation (next-Gen) sequencing–based whole-genome surveys to explore the mechanism(s) underlying such changes. In mammalian cells, cis-dependent epigenetic states are maintained by both chromatin structure and DNA methylation. Chromatin states are measured with respect to histone acetylation and methylation patterns, along with the topologies of these patterns as they extend over domains that typically encompass one or more genes. With the advent of next-generation sequencing, DNA methylation may now be characterized at the whole-genome level (MeDIP-seq) as well as at nucleotide resolution over gene-size domains (for the latter, we have optimized the rapid design and testing of tiled, nested primer pairs suitable for bisulfite-modified DNA). Of particular interest are interactions between the chromatin- and DNA methylation–based components of epigenetic control.
Given the rapid progress of epigenomics and the particularly large data sets generated by chromatin immunoprecipitation (ChIP) combined with next-Gen sequencing–based (ChIP-seq) analyses, ongoing refinement of bioinformatics tools is essential. Accordingly, we constantly improve and extend our genome annotation, pattern recognition, and pattern comparison algorithms. For topology-based mapping of chromatin patterns, we are collaborating with the Section on Chromatin and Gene Expression (headed by David Clark), who studies the small yeast genome, which permits much deeper sequencing coverage and thus much higher resolution patterns. Finally, new bioinformatic tool development will permit efficient linkage of patterns in RNA expression data sets with epigenome features.
To date, chromatin-related results indicate that genes subject to both differentiation and developmental controls depend on the three-dimensional topology of the genome and are sensitive to the remodeling of higher-order chromatin structures. We will continue to focus on large (on the order of 100 kb) domains over which histone acetylation or histone H3-K27 patterns are altered, as well as on control elements that may assume non–B DNA conformations. DNA methylation–related results suggest that a little-studied subcompartment of the genome consisting of small sets of regions with high homology, within either gene clusters or dispersed gene families, may be substantially enriched for post-natal developmental– and age-related epigenome remodeling. If our hypothesis is correct, such regions may provide clock-like functions of considerable importance.
The emerging goal is to generalize our paradigm of dynamic postnatal epigenome structure to address a range of current problems in maternal reproductive health, pediatrics, and age-related disease. Based on the genes currently under study, we will most likely investigate deficiencies in the innate immune systems of newborns, peripheral insulin resistance and diabetes in adolescents and young adults, and a spectrum of neurodegenerative processes, including Parkinson's and Alzheimer's diseases, in the elderly.
Additional Funding
- Supplementary funding from the Program in Reproductive and Adult Endocrinology, NICHD
Publications
- Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc Natl Acad Sci USA 2011;108:12687-12692.
- Cole HA, Howard BH, Clark DJ. Activation-induced disruption of nucleosome position clusters on the coding regions of Gcn4-dependent genes extends into neighbouring genes. Nucleic Acids Res 2011;[Epub ahead of print].
Collaborators
- David J. Clark, PhD, Program in Genomics of Differentiation, NICHD, Bethesda, MD
- Jonathan Epstein, MS, Molecular Genomics Laboratory, NICHD, Bethesda, MD
- Roberto Romero, MD, Program in Perinatal Research and Obstetrics, NICHD, Detroit, MI
Contact
For more information, email howard@helix.nih.gov.

