You are here: Home > Section on Formation of RNA
Physiological, Biochemical, and Molecular Genetic Events of Recognition and Resolution of RNA/DNA Hybrids

- Robert J. Crouch, PhD, Head, Section on Formation of RNA
- Susana M. Cerritelli, PhD, Staff Scientist
- Hyongi Chon, PhD, Postdoctoral Fellow
- Lina Gugliotti, PhD, Postdoctoral Fellow
- John B. Holmes, BS, Predoctoral Fellow, NIH/Oxford/Cambridge Graduate Partnerships Program
- Mariya London, BS, Technical IRTA
- Kiran Sakhuja, MS, MSc, Research Assistant
Damaged DNA is one of the leading causes of many human diseases and disorders. We study the formation and resolution of RNA/DNA hybrids, which occur during DNA and RNA synthesis. Such hybrid molecules may lead to increased DNA damage but may also play critical roles in normal cellular processes. We are interested in how RNA/DNA hybrids are resolved and in the role that ribonucleases H (RNases H) play in their elimination. Two classes of RNases H are present in most organisms. Our studies have shown that mice deleted for the Rnaseh1 gene arrest embryonic development at day 10 as a result of failure to amplify mitochondrial DNA. Others have found that the Aicardi-Goutières Syndrome (AGS), a severe neurological disorder with symptoms appearing at or soon after birth, can be caused by defective human RNase H2. We employ molecular-genetic and biochemical tools and yeast and mouse models in our research.
Click image to enlarge.
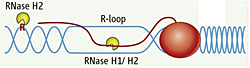
Figure 1. RNA/DNA hybrids and single rNMPs in DNA are substrates for RNase H2.
DNA is in blue, RNA is red line and letter R. The red oval represents RNA polymerase.
RNA/DNA hybrids can be composed of one strand of RNA and a second strand of DNA or can be in the form of an R-loop in which the two DNA strands are separated, with only one hybridized to RNA while the other is single-stranded DNA. A third "hybrid" is a single ribonucleotide in duplex DNA. The first two types of hybrid are substrates for class I and II RNases H. The third is uniquely recognized by type 2 RNases H (Figure 1).
Simple (non-R-loop) RNA/DNA hybrids are formed when the HIV-AIDS virus copies its genomic RNA into DNA using a reverse transcriptase (RT). The RT also has an RNase H domain that is structurally and functionally similar to the class I cellular RNase H and is necessary in several steps of viral DNA synthesis. It is known that, during RNA synthesis, R-loops can form and that aberrant R-loop formation may result in chromosome breakage. However, R-loop formation has been observed in the normal recombination process of switching (recombination) from one form of immunoglobulin to another, resulting in different isoforms of antibodies. Single-ribonucleotide hybrids occur during DNA replication, and the ribonucleotide must be removed, a process initiated by RNase H2.
Contrasts between Class I and II RNases H
Click image to enlarge.
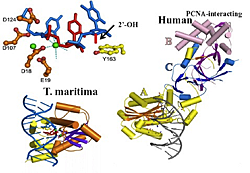
Figure 2. Structures of T. maritima and human RNases H2 complexed with substrate
Top left: Single ribonucleotide (red) in DNA (blue). Catalytic residues (brown). Metal ions (green). Y163 (yellow) pushes DNA up. Short red bar is 2′-OH conflict with Tyr. Only one strand of duplex substrate is shown. Bottom left: complex of T. maritima RNase H2 with nucleic acid. Right: Human RNase H2 modeled with T. maritima substrate. A subunit in yellow, B subunit in violet, C subunit in blue.
Previous Annual Reports have detailed our work on RNase H1. RNase H1 recognizes the 2′-OH of four consecutive ribonucleotides while the DNA strand is distorted to fit in a pocket of the enzyme. Thus, the enzyme requires more than one ribonucleotide for cleavage of RNA in RNA/DNA hybrids. In both eukaryotes and prokaryotes, RNases H1 consist of a single polypeptide. In contrast, RNase H2 is a complex of three different polypeptides in eukaryotes but a single polypeptide in prokaryotes. One of the subunits of the heterotrimeric RNase H2 of eukaryotes is similar in primary amino acid sequence to the prokaryotic enzyme. RNase H2 can recognize and cleave a single ribonucleotide or the transition from the ribonucleotide in the case of RNA-primed DNA synthesis (e.g., rrrrrDDDD in DNA—italics indicate transition from ribonucleotide to deoxyribonucleotide). In collaboration with Marcin Nowotny, we determined the three-dimensional structures of a bacterial RNase H2 from Thermotoga maritima in complex with a duplex DNA containing a single ribonucleotide. We also obtained the crystal structure of human RNase H2 in the absence of substrate (the apo form). T. maritima RNase H2 has a strong preference for ribo- to deoxyribose transition substrates and cleaves other RNA/DNA hybrids inefficiently. Our findings demonstrate an interesting mechanism for this type of hydrolysis whereby the deoxyribonucleotide downstream of the ribonucleotide is pushed up, allowing the phosphate between the ribo- and deoxyribose to interact with a Mg2+ employed in catalysis (Figure 2). In type 1 RNases H, the divalent metal ions required for cleavage are coordinated only by amino acids.
The apo structure of human RNase H2 shows that the protein subunits are intertwined, particularly for the B and C subunits (Figure 2). Subunit A is structurally similar to T. maritima RNase H2, and we were able to model the nucleic acid structure of the T. maritima structure on the human A subunit. The model predicts contacts between the nucleic acid and the enzyme. To confirm the model, we altered several of those amino acids and, as predicted by the model, enzymatic activity was reduced. Subunit B has the ability to interact with proliferating cell nuclear antigen (PCNA), a protein important in DNA replication and repair. However, the B subunit is quite distant from the catalytic center, and how PCNA and RNase H2 interact with respect to substrates remains an interesting question. Of even more interest is the question of the function of the B and C subunits other than to contribute stability to the A subunit for catalysis, in particular, whether the subunits contribute to cleaving RNA/DNA hybrids in addition to single ribonucleotide substrates.
AGS can result from mutations in RNase H2: 29 different mutations in RNase H2 are associated with AGS: eight in the A subunit, 14 in the B subunit, and seven in the small C subunit. We previously expressed, in E. coli, human RNase H2 bearing various mutations seen in AGS patients' RNases H2 and found only one with significant loss of RNase H2 activity. The structure we determined allows us to locate all known mutations in RNase H2 causing AGS. The wide distribution of these mutations suggests that modest changes in stability and interaction with other unknown proteins and modest reductions in catalysis can all cause AGS.
The difference in substrates cleaved by bacterial and eukaryotic RNases H2 in vitro raises the question as to the mechanism underlying the expanded repertoire of the eukaryotic enzymes. The interaction of the Tyr residue with the deoxyribose in T. maritima RNase H2 would not be possible were a ribose present due to a conflict with the 2′-OH of the ribose. One possibility for expanded substrate recognition is that the conserved Tyr is more flexible in eukaryotic RNases H2 than in the bacterial enzymes. This is an important question because single-ribonucleotide substrates appear only during DNA replication and repair whereas R-loop structures can occur any time that transcription takes place.
In collaboration with Ian Holt, we are searching for possible roles of RNase H1 in mitochondrial DNA replication by using, among other techniques, analysis of intermediates on two-dimensional gels. Our findings thus far indicate that elevated expression of RNase H1 in mitochondria alters mtDNA replication. In addition, data obtained in Holt's laboratory by John Holmes supporting the existence of RNA/DNA hybrids as replication intermediates indicate a bi-directional mode of DNA replication for this organelle.
Publications
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes from eukaryotes. FEBS J 2009;276:1494-1505.
- Rychlik MP, Chon H, Cerritelli SM, Klimek P, Crouch RJ, Nowotny, M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol Cell 2010;40:658-670.
- Figiel M, Chon H, Cerritelli SM, Cybulska M, Crouch RJ, Nowotny M. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutières Syndrome defects. J Biol Chem 2011;286:10540-10550.
- Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM. Contributions of the two accesory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res 2009;37:96-110.
- Cerritelli SM, Chon H, Crouch, RJ. A new twist for topoisomerase. Science 2011;332:1510-1511.
Collaborators
- Peter Burgers, PhD, Washington University, St. Louis, MO
- Ian Holt, PhD, MRC-Dunn Nutrition Unit, Cambridge, UK
- Shigenori Kanaya, PhD, Osaka University, Osaka, Japan
- Paul E. Love, MD, PhD, Program on Genomics of Differentiation, NICHD, Bethesda, MD
- Herbert C. Morse, MD, Laboratory of Immunopathology, NIAID, Bethesda, MD
- Marcin Nowotny, PhD, International Institute of Molecular and Cell Biology, Warsaw, Poland
Contact
For more information, email crouch@helix.nih.gov or visit sfr.nichd.nih.gov.

