You are here: Home > Section on Vertebrate Development
Control of Ectodermal Development in Vertebrate Embryos

- Thomas D. Sargent, PhD, Head, Section on Vertebrate Development
- Hiu Wan Law, Visiting Fellow
- Yoko Ogawa, PhD, Visiting Fellow
- Valerie Virta, PhD, Intramural Research Training Award Fellow
The primary interest of this group is the control of cranial neural crest (CNC) development. We started the project by identifying the transcription factor TFAP2 as a critical component of NC induction in the frog Xenopus, then went on to study regulatory targets of TFAP2, including a novel protein we named Inka. Inka acts as a regulator of cytoskeletal dynamics, partly by interacting with a protein kinase, Pak4. The kinase is known to mediate cell-cell signals that influence cell shape, adhesion, and migration, both in development and in cancer. The focus of the Inka project has shifted to Pak4 function in zebrafish development in the past year. We have also initiated two new CNC projects, also being carried out in zebrafish: one is to use the mutation dlx3TDO in the homeobox gene dlx3. This dominant mutation is the basis for a human genetic disorder affecting bones, teeth, and hair, and converts the transcription factor into a dominant-negative inhibitor of wild-type Dlx3. We plan to probe the function of Dlx3 in zebrafish using this molecule as a tool. The other project is to probe, and then to manipulate, cell-cell signaling in migrating CNC cells in the living embryo, basing our work on transgenic expression in CNC cells of a fluorescence resonance energy transfer (FRET) biosensor, followed by regulated expression of engineered signaling molecules.
Pak4 function in zebrafish
Click image to enlarge.
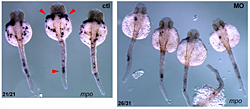
Figure 1. Pak4 knockdown inhibits neutrophil development.
Knockdown of zebrafish Pak4 by morpholino oligonucleotides drastically reduces the formation of neutrophils. Visualized by in situ hybridization with a probe for myeloperoxidase (mpo; red arrowheads), a neutrophil-specific gene.
PAK4 (p21-activated kinase 4) belongs to a family of six genes in human. PAKs bind to small GTPase effector molecules, the Rho-class proteins Cdc42 and Rac, and transduce signals to the cytoskeleton and nucleus, affecting cell shape and gene expression, respectively. Overexpression of Pak4 is tumorigenic in mouse cells, and the abundance of PAK4 in many tumors suggests this gene mediates a common step in cancer progression. In the mouse, disruption of the Pak4 gene yields a complex embryonic lethal phenotype affecting vasculature, heart, and neuronal and other tissues. Our work in zebrafish showed that pak4 in this species is most strongly expressed in the egg, i.e., maternally, and it is this maternal expression that is essential while the zygotic pak4 can be dispensed with. Loss of maternal pak4 in zebrafish results in a complex lethal phenotype, including abnormal axial muscle development and defects in hematopoiesis, most strikingly, the suppression of neutrophils, cells involved in inflammatory response to infection or injury. Other tissues, including the cranio-facial cartilage, were also affected in the knockdown embryos.
The next phase of this project will be to elucidate the regulatory pathways that are affected by loss of maternal Pak4. Genomic strategies such as massive RNA sequencing will be used to identify targets for altered gene expression. We also plan to carry out a yeast two-hybrid screen to search for novel Pak4 interactions. As candidate genes and/or proteins are identified, we can use the Pak4 loss-of-function phenotype as a standard for comparison for future loss-of-function experiments with these candidates. We will focus on the hematopoiesis and somite aspects of the Pak4 phenotype, but will also monitor other aspects, particularly cranio-facial development.
Dlx gene function in cranial neural crest development
Click image to enlarge.
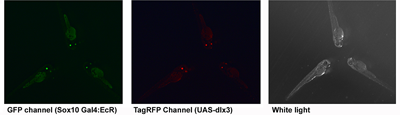
Figure 2. Transgenic zebrafish
Double-transgenic zebrafish produced using Tol2 recombinase. Transgenesis markers are eGFP and TagRFP driven by a zebrafish alpha crystallin promoter (lens). About 100 independent founders have been made.
The distal-less class of homeobox genes have been linked for some time with the regulation of cranio-facial morphology. Of this group of six genes (Dlx1-6 in mammals) Dlx1, Dlx2, Dlx5, and Dlx6 have been most extensively investigated in this context. We showed in Xenopus that dlx3 expression is excluded from CNC at early stages, that this could be due to differential sensitivity to local BMP signal strength, and that ectopic expression of dlx3 (but not other dlx genes) in the early CNC obliterated this differentiation pathway. Knockdown of dlx3b in zebrafish predominantly leads to defects in the otic (ear) placode. However, combined knockdown of dlx3b and other dlx genes results in multiple jaw defects, indicating redundant functions. In humans, the dominant autosomal genetic disorder tricho-dento-osseous syndrome (TDO) affects some aspects of CNC development, and it is thought that this occurs by a dominant-negative effect on the wild-type DLX3 allele, or possibly on other members of this superfamily (DlX and related MSX factors can form heterodimers in vivo). We plan to use this naturally occurring mutant dlx3 to inhibit the wild-type Dlx3 protein, and potentially other co-expressed dlx genes, in the zebrafish CNC. It is important to have spatial and temporal control of expression for this approach. To achieve this, we plan to use a hormone (ecdysone)-inducible fusion of a modified Gal4 transcriptional activator, driven in transgenic zebrafish embryos with the promoter of the sox10 gene, which is neural crest–specific at early stages of development. This will be used in conjunction with zebrafish carrying a transgene with an upstream activating sequence (UAS) linked to the human DLX3TDO gene. As a shortcut, we have been introducing both transgenes together, via the Tol2 recombinase method. Transgenesis is detected using a GFP or RFP reporter driven by a zebrafish alpha crystallin promoter. This gives green or red eyes three days post fertilization or yellow eyes if both are expressed. We find that about 95% of fish that express one transgenesis marker also express the other, suggesting that four to five copies have been integrated per diploid genome. When mature, double-transgenic fish will be outcrossed to non-transgenic mates. At least 1/4 of the offspring embryos should also have both genes and can be tested for target gene expression by treatment with the ecdysone inducer (tebufinozide) to identify high-efficiency regulated expressing lines. These should enable us to identify the critical period of dlx gene function for various elements in the craniofacial skeleton, and perhaps also in the pharygeal dentition.
In addition, we hypothesize that the exclusion of Dlx3 from early CNC will also prove to be essential in zebrafish, as it is in Xenopus. To test this, we will drive zebrafish dlx3b expression in premigratory CNC using the binary system we are working out for the dominant-negative strategy described above. Our findings will be interpreted in the light of equivalent experiments being carried out in mouse via gene replacement by our collaborator Maria Morasso.
Imaging signal transduction in live neural crest cells
Click image to enlarge.
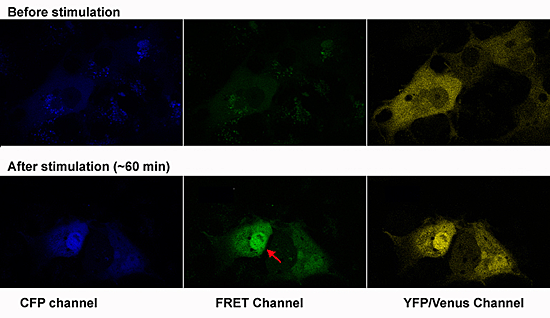
Figure 3. Testing a FRET biosensor
Test of the FRET MAPK biosensor in Cos7 cells. Transfected cells were starved for 16 hr then induced with 10% FBS for 30 minutes. The CFP-MAPK-YFP fusion can be seen translocating to the nucleus (red arrow), accompanied by increased FRET.
Neural crest cells acquire much of their identity by inductive interactions with adjacent tissues during migration. CNC cells interact with the mesodermal phyaryngeal pouches into which they migrate, and with overlying ectoderm. Such interactions are essential for proper cranio-facial morphogenesis and differentiation. The goal of this project is to visualize these cell-cell interactions in living embryos by monitoring signal transduction components using FRET. We have adapted a biosensor, MIU2 (http://www.lif.kyoto-u.ac.jp/labs/fret/e-phogemon/phomane.htm) so that it can be expressed specifically in the neural crest of transgenic zebrafish. When FGF or other signal pathways that activate p42 MAPK become stimulated, the FRET signal should increase, allowing us to see this in the embryo; we will be using wild-type, mutant, and "morphant" zebrafish for such experiments. Ultimately, we plan to use the ecdysone-inducible Gal4 binary expression system described in the previous section to drive expression of dominant-negative or constitutively active signal transduction intermediates at variable times during CNC migration, enabling us to test hypotheses regarding specific pathways. Once the tools have been developed for monitoring MAPK signaling, we can adapt them to other signal transduction pathways mediated by other effectors. Moreover, we can use these tools to probe the downstream cellular changes that occur as the CNC cells differentiate into cranio-facial cartilage elements.
The Inka project
Last year, we and our collaborators completed analysis of knockout mice lacking the Inka1 gene. While Inka-null embryos had a higher than expected incidence of cranio-synostosis (about 5%), the vast majority developed normally into apparently healthy, fertile adults. This year, we completed a parallel loss-of-function project in zebrafish. We crossed nonsense-truncated inka1a alleles, identified by the TILLING (targeting induced local lesions in genome) method, yielding homozygous F2 animals, which invariably developed normally. This was done with two different mutant alleles, and in three different zebrafish background strains, with the same result. We concluded that inka1a function is less critical than we were led to believe from our early morpholino knockdown experiments. While inka overexpression has interesting effects on cytoskeletal structure, cell adhesion, and morphogenesis, the absence of a loss-of-function phenotype makes further research on the embryonic function of inka1 much more difficult, so this project has been put on hold.
Publications
- Reid BS, Sargent TD, Williams T. Generation and characterization of a novel neural crest marker allele, Inka1-LacZ, reveals a role for Inka1 in mouse neural tube closure. Dev Dyn 2010;239:1188-1196.
- Hwang Y-S, Luo T, Xu Y, Sargent TD. Myosin-X is required for cranial neural crest cell migration in Xenopus laevis. Dev Dyn 2009;238:2522-2529.
Collaborators
- Maria Morasso, PhD, Laboratory of Skin Biology, NIAMS, Bethesda, MD

