You are here: Home > Section on Eukaryotic Gene Regulation
Regulation of Mammalian Cell Proliferation and Development

- Melvin L. DePamphilis, PhD, Head, Section on Eukaryotic Gene Regulation
- Christelle De Renty, PhD, Visiting Fellow
- Kotaro Kaneko, PhD, Staff Scientist
- Chrissie Lee, PhD, Postdoctoral Fellow
- Haiyan Pan, PhD, Postdoctoral Fellow
- Zakir Ullah, PhD, Visiting Fellow
- Alex Vassilev, PhD, Staff Scientist
- Xiaohong Zhang, BA, Technical Assistant

Click image to enlarge.
Figure 1. Genome Duplication (Garland Science, 2010) by Melvin L. DePamphilis and Stephen D. Bell
This is the first comprehensive description of the subject since Kornberg and Baker's book DNA Replication, second edition (W.H. Freeman, 1992). Genome Duplication describes the principles, concepts, and mechanisms common to genome duplication in all three domains of life: bacteria, archaea, and eukarya. In addition, it discusses the evolution of the DNA replication machinery and the impact of DNA replication on human disease.
Nothing is more fundamental to living organisms than the ability to reproduce. Each time a human cell divides, it must duplicate its genome, a problem of biblical proportions. A single fertilized human egg contains 2.1 meters of DNA. An adult of about 75 kg (165 lb) consists of about 29 trillion cells containing a total of about 60 trillion meters of DNA, a distance equal to 400 times the distance from the Earth to the sun. Not only must the genome be duplicated trillions of times during human development, but it must be duplicated once and only once each time a cell divides (termed mitotic cell cycles). If we interfere with this process by artificially inducing cells to re-replicate their nuclear genome before cell division, the result is DNA damage, mitotic catastrophe, and programmed cell death (apoptosis). On rare occasions, specialized cells can duplicate their genome multiple times without undergoing cell division (termed endocycles), but when this occurs, it generally results in terminally differentiated polyploid cells that are viable but no longer proliferate. As we age, however, the ability to regulate genome duplication diminishes, resulting in genome instability, which allows genetic alterations that can result in promiscuous cell division—better known as cancer. For a comprehensive description of genome duplication in all forms of life, refer to 'Genome Duplication' (Figure 1).
Our research program focuses on three questions: What are the mechanisms that restrict genome duplication to once per cell division? How are these mechanisms circumvented to allow developmentally programmed induction of polyploidy in terminally differentiated cells? How can we manipulate these mechanisms to destroy cancer cells selectively?
Recent accomplishments
Anti-geminin drugs. Previously, we discovered that suppression of geminin, a protein that restricts genome duplication to once per cell division, induced DNA re-replication and apoptosis in cells derived from human cancers without harming cells derived from normal human tissues (U.S. Patent Application pending), suggesting that anti-geminin compounds could provide a novel chemotherapy (1). To this end, we developed a high-throughput screening assay to search for anti-geminin drugs that may be useful in cancer chemotherapy and for basic research on genome duplication and cell proliferation (2). We are also using this assay to screen siRNA libraries of the human genome for genes essential for restricting genome duplication in cancer cells, but not in normal cells, because normal cells have additional mechanisms in place that regulate genome duplication.
ORC assembly. The origin recognition complex (ORC) consists of six proteins that are essential for licensing DNA replication origins. We discovered that Orc6, Orc1, and ORC(2–5) are transported independently to the nucleus, where they can either assemble into ORC(1–6) or function individually (3). This discovery accounts for the fact that various ORC subunits appear to play a role in heterochromatin assembly, ribosomal biogenesis, centrosome and kinetochore function, sister chromatin cohesion, and neural dendritic branching. Collectively, these varied and seemingly contradictory observations suggest that ORC assembly is a dynamic rather than static process.
Checkpoint kinase-1. Trophoblast stem (TS) cells are unique in that they differentiate into the trophoblast giant (TG) cells required for implantation and placentation. We previously reported that endoreduplication in TS cells is triggered by p57 inhibition of CDK1 with concomitant suppression of the DNA damage response by p21. We recently discovered a novel role for checkpoint kinase-1 (CHK1). In addition to its essential function in the DNA–damage response in mammalian cells, CHK1 is part of a "G2 restriction point" that prevents premature cell cycle exit in cells programmed for terminal differentiation, a role that CHK2 cannot play (4). We are currently investigating the features of CHK1 that are unique to each role and the role p21 plays in suppressing the DNA damage response in TG cells (see also next section).
Role of TEAD4 in regulating energy homeostasis. We previously reported that the transcription factor TEAD4 specifies the trophoectoderm (TE) lineage at the beginning of mammalian development; to date, TEAD4 remains the earliest zygotically expressed transcription factor essential for differentiation of totipotent cells into the TE lineage. However, we recently discovered that TEAD4 is actually required for regulating energy homeostasis during pre-implantation development, a role that is essential in order for other genes to specify the trophectoderm lineage (submitted for publication).
SoxC interaction with TEAD2. SoxC transcription factors play a critical role in organogenesis. Specific inactivation of SoxC genes in neural and mesenchymal cells leads to selective apoptosis of these cells. TEAD2 functionally interacts with SoxC genes in embryonic development and is a direct target of SoxC proteins (5). SoxC genes therefore ensure neural and mesenchymal progenitor cell survival and function in part by activating this transcriptional mediator of the Hippo signaling pathway. Whether TEAD2, like TEAD4, regulates energy homeostasis remains to be determined.
Regulation of DNA replication in mammalian cells
Click image to enlarge.
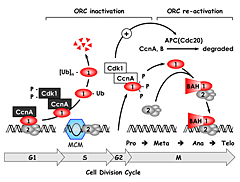
Figure 2. The ORC cycle in mammalian cells (see Noguchi et al., EMBO J 2006;25:5372 and references therein)
ORC(1–6) is bound to chromatin during the G1 phase of the cell cycle, at which time it is part of a prereplication complex. When S phase begins, the association between Orc1 and chromatin-bound ORC(2–6) is destabilized by selective CDK–dependent phosphorylation and ubiquitination. Monoubiquitinated Orc1 is exported to the cytoplasm. The remaining ORC subunits are subsequently released from chromatin. Polyubiquitinated Orc1 is degraded by the 26S proteasome. Orc1 levels are restored during the G2-to-M transition, but Orc1 is hyperphosphorylated, an event that prevents ORC assembly. During the anaphase-to-G1 phase transition, Orc1 is dephosphorylated and, together with other ORC subunits, binds to chromatin, an event facilitated by the Orc1 BAH domain. If Orc1 is not associated with other ORC subunits, or if it is not phosphorylated or ubiquitinated, then it induces apoptosis.
DNA replication begins when the six-subunit ORC binds to specific chromosomal loci termed origins of bidirectional replication, which we and others mapped at specific sites in the genomes of flies and mammals. These sites are determined by both genetic and epigenetic features. The number and locations of replication origins in the cells of multicellular organisms can change from an average of one every 10 to 20 kb in the rapidly cleaving embryos of frogs, flies, and fish to one every 50 to 300 kb in the differentiated cells of adult organisms. Developmental changes in origin density also occur during specific stages in animal development. Thus, metazoan genomes contain many potential replication origins, but during development, some of these sites are selectively activated while others are suppressed, a concept introduced many years ago as the "Jesuit Model" (many are called, but few are chosen) (M.L. DePamphilis, Ann Rev Biochem 1993;62:29; Curr Opin Cell Biol 1993;5:434; J Biol Chem 1993;268:1).
ORC initiates assembly of prereplication complexes (preRCs) consisting of a DNA helicase loader [ORC(1–6), Cdc6, and Cdt1] and the replicative DNA helicase [Mcm(2–7)]. Several years ago, we discovered that the behavior of ORC in mammalian cells differs significantly from that in single-cell eukaryotes such as yeast. In contrast to yeast, Orc1 associates weakly with the stable core complex ORC(2–5), and the ability of ORC to initiate DNA replication depends on this interaction. Moreover, it appears that the interaction of Orc1 with ORC(2–5) is one of the mechanisms that regulates when and where initiation events occur. We termed this concept the ORC cycle, and we and others have established its basic features (Figure 2). Cell cycle–dependent modifications of Orc1 regulate Orc1 activity, and Orc1 activity regulates ORC activity, which regulates initiation of DNA replication.
Click image to enlarge.
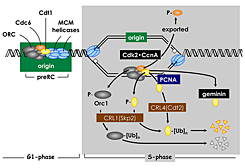
Figure 3. Multiple convergent pathways restrict genome duplication to once per cell division.
In human cells, Cdt1 activity is down-regulated in four ways. Free Cdt1 is phosphorylated by Cdk2 in association with Cyclin A (CcnA), thereby suppressing Cdt1 activity and converting Cdt1 into a substrate for the CRL1(Skp2) ubiquitin ligase. As replication forks pass through the origin, Cdt1 binds to the proliferating cell nuclear antigen (PCNA) clamp, which holds the replicative DNA polymerase onto the replication fork. This Cdt1•PCNA•chromatin form of Cdt1 is a substrate for the CRL4(Cdt2) ubiquitin ligase. Ubiquitinated Cdt1 is then degraded by the 26S proteasome. Finally, geminin binds to Cdt1 and inhibits its activity. In addition to inactivation of Cdt1, CcnA–dependent phosphorylation of Orc1 prevents it from binding to chromatin during mitosis, and CcnA–dependent phosphorylation of Cdc6 suppresses its activity and promotes its nuclear export.
The ORC cycle is only one of six known mechanisms that can determine when and where DNA replication begins in human cells. Cdk2•CcnA also suppresses Cdc6 and Cdt1 activities by phosphorylation, and Cdt1 is targeted by two ubiquitin ligases and by geminin, a Cdt1–specific protein inhibitor (Figure 3). However, not every pathway is active in all cell types. For example, cells derived from human cancers are dependent on geminin to prevent DNA re-replication, whereas cells derived from normal human tissues are dependent on geminin as well as on cyclin A–dependent CDK activity.
A rare event in mammals, endoreduplication, by which the nuclear portion of the genome is duplicated one or more times (endocycles) without an intervening mitosis, is a common event among arthropods and plants. In mammals, it first occurs during peri-implantation development when cells within the TE of the blastocyst are deprived of the mitogenic factor FGF4. TS cells then differentiate into the polyploid, viable, nonproliferating TG cells required for embryo implantation and placentation (Figure 4). PreRC assembly requires the absence of both CDK activity and geminin, a condition that occurs in mitotic cell cycles during the anaphase-to-G1 phase transition. We discovered that this condition could be induced in TS cells by selective chemical inhibition of CDK1, the enzyme required for entry into mitosis, but not in ES cells, in which the consequence is apoptosis. Therefore, selective inhibition of CDK1 triggers endoreduplication only in cells programmed to differentiate into polyploid cells. Similarly, FGF4-deprivation of TS cells induced expression of two CDK–specific inhibitors: p57/Kip2 and p21/Cip1. One of them (p57) was essential for endoreduplication, and the other (p21) appeared to facilitate p57 activity by suppressing expression of Chk1 and Emi1 (Figure 5). TS cells (+FGF4) express both the p57 and p21 genes, but the nonactivated form of Chk1 phosphorylates the p57 and p21 proteins, thereby targeting them for ubiquitin-dependent degradation. In TG cells, CHK1 is suppressed to allow p57 expression during G phase, CDK2 is required for DNA replication, and p57 is degraded during S phase to allow endocycles.
DNA replication and cell differentiation during pre-implantation development
Click image to enlarge.
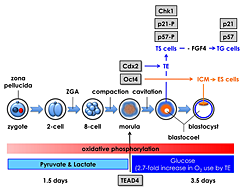
Figure 4. Mouse pre-implantation development; zygotic gene activation (ZGA) begins at the two-cell stage.
The eight totipotent blastomeres at the eight-cell stage compact into a morula, and transcription factors Tead4, Cdx2, and Oct4 mark the beginning of a chain of events that specifies trophectoderm (TE) and inner cell mass (ICM). Oct4 is the first gene whose expression is essential for maintaining the blastomeres in a totipotent state, thereby producing the ICM and embryonic stem (ES) cells that differentiate into embryonic tissues. Tead4 is the first gene whose expression is essential for differentiation of blastomeres into TE and trophoblast stem (TS) cells that produce the placenta. In the absence of Tead4 expression, all totipotent blastomeres produce OCT4 protein, revealing that TEAD4 protein is required for Cdx2 gene expression. However, TEAD4 is essential for blastocyst formation only under conditions that increase the rate of oxidative phosphorylation. Following compaction, pre-implantation embryos switch energy substrates from pyruvate and lactate to glucose in order to meet the increased energy demands of blastocoel formation, an event unique to the TE. Thus, Tead4 does not specify TE; TEAD4 maintains energy homeostasis so that other genes can specify TE.
Fertilization activates the first round of genome duplication, after which the fertilized egg cleaves into a two-cell embryo (Figure 4). In mice, the two-cell embryo then activates expression of about 300 genes that are required to continue development of the organism (termed zygotic gene activation). Initially, every cell (blastomere) produced by the cleavage events is "totipotent"; that is, it is capable of giving rise to the entire organism. But, within five rounds of cell cleavage, a blastocyst appears, marking the beginning of cell differentiation. Blastocysts consist of a spherical monolayer of epithelial cells called the TE that gives rise to TS cells, TG cells, and eventually to the placenta. The TE layer encompasses a group of cells called the inner cell mass that gives rise to embryonic stem (ES) cells and eventually to the embryo. Our goal for some years has been to determine whether the requirements for genome duplication in cultured cells were the same for cleavage-stage embryos, for the stage prior to cell differentiation, and for the transition from mitotic cell cycles to endocycles.
Click image to enlarge.
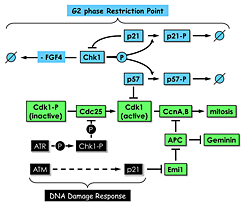
Figure 5. Proliferating TS cells exhibit a G2 phase restriction point analogous to the G1 restriction point in proliferating cultured mammalian cells.
In the presence of the mitogen FGF4 and in the absence of DNA damage, Chk1, the same checkpoint kinase that is activated by DNA damage, phosphorylates both p57 and p21, thereby targeting them for ubiquitin-dependent degradation. In the absence of FGF4 and DNA damage, Chk1 expression is suppressed by an as yet unidentified mechanism. Both p57 and p21 proteins accumulate. P57 inhibits CDK1, and TS cells exit their mitotic cell cycle and differentiate into trophoblast giant (TG) cells. P21 play several roles, including suppressing Chk1 expression to maintain TG cell status and inhibiting Emi1 to activate the anaphase promoting complex (APC), which promotes preRC assembly by targeting cyclins A and B and geminin for degradation. Both Chk1 and p21 are also components of the DNA damage response that prevents mitosis until the problem is corrected. The features that distinguish their roles in these two regulatory mechanisms remain to be elucidated.
To this end, we identified genes that we thought might be critical for pre-implantation development. The results have been fruitful, but surprising. We discovered that Dkkl1, a gene unique to mammals, is expressed specifically during implantation of the embryo and development of spermatocytes into sperm. Moreover, we showed that inactivation of Dkkl1 in mice resulted in production of sperm that are defective in fertilization. We discovered that Tead2, one of a highly conserved family of four transcription factors that share a common DNA–binding domain, is expressed from the two-cell embryo throughout pre-implantation development but remarkably is not required until after implantation and the start of nervous system formation. Mice lacking a functional Tead2 gene have difficulty forming a neural tube. Failure to close the neural tube in mice is called exencephaly, which is related to anencephaly, a common human birth defect that can be prevented by folic acid. Finally, we discovered that Tead2 and Tead4 are the only Tead genes expressed in pre-implantation mouse embryos and that, in contrast to genetic inactivation of Tead2, genetic inactivation of Tead4 resulted in formation of morphologically abnormal morula, the absence of TE-specific genes, and failure to produce a blastocoel. These and other results in the literature strongly suggested that Tead4 is a master gene that sets in motion the first round of cell differentiation during mammalian development. Surprisingly, our subsequent research demonstrated otherwise.
Master genes trigger a sequence of events that specify cell fate. However, genes essential for maintaining a metabolic state that allows other genes to specify cell fate may be mistaken for master genes. We have validated this concept by showing that Tead4, the presumptive master gene for trophectoderm specification, is a homeostatic gene that is essential for blastocoel formation in utero, a prerequisite to embryo implantation and placentation. Requirement for Tead4 is induced metabolically, not developmentally, and conditions that minimize oxidative stress allow embryos lacking TEAD4 to develop into blastocysts expressing genes required to produce functional trophectoderm. TEAD4, which is expressed at the eight-cell stage, is essential only under conditions that promote energy production by oxidative phosphorylation (Figure 4). Given that either inhibition of the mTOR pathway or an anti-oxidant alleviates this requirement, TEAD4 appears to regulate changes in redox potential that are expected to occur during high levels of oxidative phosphorylation associated with blastocyst formation in utero.
Additional Funding
- Director's Challenge Award for siRNA screening of human genome
Publications
- Depamphilis ML. Spotlight on geminin. Breast Cancer Res 2011;13:109.
- Zhu W, Lee CY, Johnson RL, Wichterman J, Huang R, DePamphilis ML. An image-based, high-throughput screening assay for molecules that induce excess DNA replication in human cancer cells. Mol Cancer Res 2011;9:294-310.
- Ghosh G, Vassilev AP, Zhang J, Zhao Y, DePamphilis ML. Assembly of the human origin recognition complex occurs through independent nuclear localization of its components. J Biol Chem 2011;286:23831-23841.
- Ullah Z, de Renty C, DePamphilis ML. Checkpoint kinase 1 prevents cell cycle exit linked to terminal cell differentiation. Mol Cell Biol 2011;31:4129-4143.
- Bhattaram P, Penzo-Mendez A, Sock E, Colmenares C, Kaneko KJ, Vassilev A, Depamphilis ML, Wegner M, Lefebvre V. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun 2010;1:9-15.
Collaborators
- Stephen D. Bell, PhD, Oxford University, Oxford, UK
- Marc Ferrer-Alegre, PhD, NIH Chemical Genomics Center, NHGRI, Rockville, MD
- Rajarshi Guha, PhD, NIH Chemical Genomics Center, NHGRI, Rockville, MD
- Ronald L. Johnson, PhD, Cancer Etiology Branch, NCI, Rockville, MD
- Véronique Lefebvre, PhD, Cleveland Clinic Lerner Research Institute, Cleveland, OH
- Scott E. Martin, PhD, NIH Chemical Genomics Center, NHGRI, Rockville, MD
- Wenge Zhu, PhD, George Washington University Medical School, Washington, DC
Contact
For more information, email depamphm@mail.nih.gov or visit depamphilislab.nichd.nih.gov.

