Neuronal Circuits Controlling Behavior: Genetic Analysis in Zebrafish

- Harold Burgess, PhD, Head, Section on Behavioral Neurogenetics
- Tripti Gupta, PhD, Staff Scientist
- Jennifer L. Sinclair, MSc, Zebrafish Technician
- Ashwin A. Bhandiwad, PhD, Postdoctoral Fellow
- Jennifer Panlilio, PhD, Postdoctoral Fellow
- Svetlana Semenova, PhD, Visiting Fellow
- Nickolas Chu, BSc, Postbaccalaureate Fellow
- Jacob Clarin, BSc, Postbaccalaureate Fellow
The Section on Behavioral Neurogenetics studies how, under diverse environmental contexts, the nervous system selects appropriate behavioral responses to sensory information in a way that best satisfies internal motivational objectives. We use the larval zebrafish as a model because its brain exhibits the basic architecture of the vertebrate brain but is much less complex than the mammalian brain. Despite the relative simplicity of their nervous system, zebrafish have a sophisticated repertoire of sensory-guided and internally driven behaviors. Furthermore, the optical clarity of the embryo facilitates visualization of individual neurons and their manipulation with genetic techniques. Behavior in larvae is innate and therefore exhibits minimal variability between fish. Subtle alterations in behavior can therefore be robustly measured, making it possible to quickly assess the contribution of identified neurons to a variety of motor behaviors.
We focus on two aspects of behavioral regulation: the neuronal mechanisms by which sensory context regulates behavioral decisions and the pathways that sustain changes in behavioral state. Neuronal connections that allow the brain to integrate sensory and internal-state information are established through genetic interactions during development, and are frequently disrupted by gene mutations associated with neuro-developmental disorders. We can therefore use discoveries about sensorimotor integration pathways to understand how human disease genes disrupt brain development. To support these objectives, we develop new genetic tools and behavioral assays to probe the nexus between neuronal function and behavior at single-cell resolution.
Neuronal pathways for auditory sensory processing
Startle responses are rapid reflexes that are triggered by sudden sensory stimuli and which help animals defend against, or escape from, potentially threatening stimuli. In both fish and mammals, startle responses are initiated by giant reticulospinal neurons in the medulla, which receive short-latency sensory input from diverse sensory modalities. Although highly stereotyped, startle responses are nevertheless modulated by sensory context and behavioral state and are therefore an excellent system in which to understand how such information is integrated for behavioral choice. In mammals, including humans, the startle response to a strong auditory stimulus can be inhibited by pre-exposure to a weak acoustic ‘prepulse’. This form of startle modulation, termed prepulse inhibition, is diminished in several neurological conditions. Our work in resolving the core neuronal pathway that mediates prepulse inhibition provides a basis to probe how gene mutations linked to neuro-developmental disorders disrupt sensory processing.
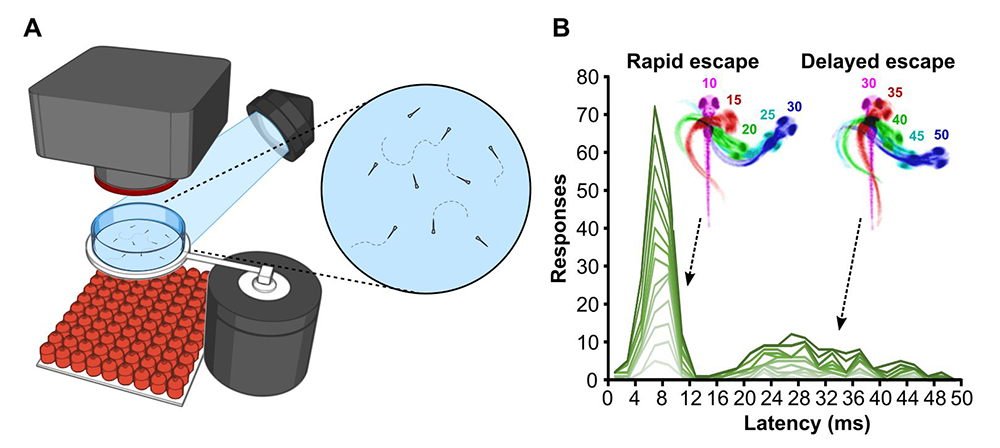
Analysis of startle control mechanisms is facilitated by using the zebrafish model. Sudden vibrational stimuli trigger rapid-escape swims in zebrafish, which are mediated by giant reticulospinal neurons, in a manner similar to the central neurons controlling startle responses in mammals. We previously demonstrated that escape swims are suppressed by pre-exposure to a prepulse, which allowed us to apply the powerful suite of genetic tools available in zebrafish to identify neurons that mediate prepulse inhibition. However, as in humans, zebrafish have more than one pathway for escape behavior. At larval stages, auditory stimuli drive either an extremely rapid Mauthner-cell mediated response, or a slightly delayed non-Mauthner behavior. Delayed responses are of special interest as they are not susceptible to prepulse inhibition; the effect of a weak prepulse is thus to increase use of delayed escapes while rapid escapes are suppressed. However, almost nothing was known about the neuronal pathway that mediates for delayed escapes. We therefore performed a circuit-breaking screen, in which we systematically ablated groups of Gal4–labeled neurons in a library of enhancer-trap lines, to identify neurons that are required to initiate delayed escape. We discovered a population of cells adjacent to the cerebellum in the prepontine hindbrain that initiate delayed escapes. Auditory cues that are insufficiently threatening to drive an immediate escape behavior activate the prepontine delayed escape population, allowing larvae to perform a flexible and graded response. The identification of such neurons provides a new paradigm for dissecting the circuit that mediates very rapid behavioral selection in an ethologically critical pathway [Reference 2].
Figure 1. Larval-stage zebrafish perform two modes of escape response distinguished by latency to initiation.
A. A high-speed camera is used to monitor behavioral responses of larvae to a short vibratory stimulus.
B. When tested over multiple trials, individual larvae (green lines) initiate escape responses in two waves: a rapid fast-swimming response, and a second, delayed response with slower movement characteristics.
Neural mechanisms for behavioral state control
Over the course of the day, motivational goals change in response to both internal and external cues. At a given moment, an individual’s behavioral state strongly influences decisions on how to interact with the environment. A major goal in neuroscience is to identify the neural systems that maintain short-term behavioral states and to determine how they interact with central mechanisms for behavioral choice. In zebrafish, loss of illumination triggers a short-term behavioral state in which larvae show heightened locomotor activity. We previously demonstrated that light-sensitive neurons in the hypothalamus trigger this a state of hyperactivity and that the response to loss of illumination is a part of a light-search behavior.
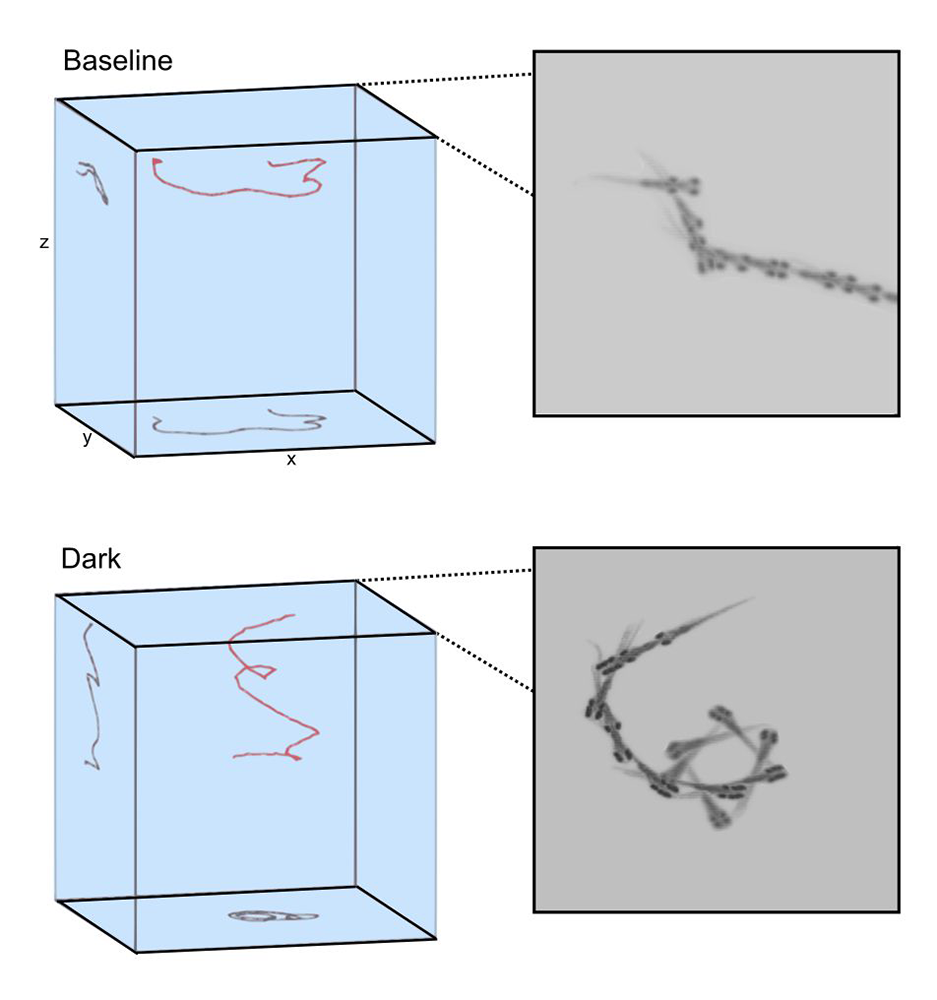
Unexpectedly, we noted that during the first two minutes in the dark, zebrafish repeatedly turn in a single direction resulting in a circular swimming pattern, and that individual fish had a tendency to circle in a left or rightward direction (Figure 2). Left/right motor identity was stable across several days, and correlated also with left/right startle preference, when larvae were tested in the absence of overriding visual cues. Motor asymmetry is common across the animal kingdom and is manifested in humans as hand-preference. Although human handedness is known to be disrupted in several neuro-developmental disorders and has been the subject of much research, little is known about how it is generated during development, or maintained within the brain. We therefore used the opportunity to leverage our observation that zebrafish have an intrinsic motor asymmetry to investigate developmental and neural mechanisms.
To achieve this, we performed a circuit-breaking screen before testing for changes in motor bias. Two lines that labeled neurons required for motor bias both included a small group of around 60 cells in the posterior tuberculum, a forebrain region ventral to the thalamus. Unilateral laser ablation of posterior tubercular neurons imposed a motor bias on treated larvae, such that larvae with an intact left posterior tuberculum tended to circle rightward, and vice versa. We also found that an output pathway running through the habenula was required for such neurons to drive motor asymmetry. The experiments define the specific neuronal basis for motor asymmetry in zebrafish.
While conducting these studies, a spontaneous mutation occurred in our wild-type stock that strongly suppressed motor asymmetry. Using a high-thoughput sequencing approach, we mapped the underlying genetic lesion to a small deletion of the first two exons in the gene epb41l5, then used an independent mutation to confirm that fish with a heterozygous mutation in this gene fail to develop normal motor asymmetry. Given that epb41l5 interacts with members of the notch signaling pathway, we also tested left/right bias in heterozygous mutants with mib mutations, and again discovered loss of motor asymmetry. The findings indicate that normal acquisition of left/right motor identity is disrupted by gene mutations that quantitatively perturb Notch signaling levels during embryonic development [Reference 3].
Figure 2. Motor asymmetry in zebrafish
After transitioning from baseline illuminated conditions to dark conditions, zebrafish slowly swim in a circular pattern toward the bottom of the arena.
Tools for decoding neuronal circuits
To enable circuit-breaking screens, we generated several hundred new Gal4 and Cre lines that can provide intersectional targeting of small groups of neurons with a high degree of specificity [References 4 & 5]. A unique feature of brain imaging in zebrafish is the ability to visualize the total architecture of the brain while simultaneously recording the position and morphology of every constituent labeled neuron. Thus, to make these tools accessible to the broader research community, we performed whole-brain imaging for each line, then registered the image of each line to the same reference brain. In collaboration with Nicholas Polys, we then developed an online brain atlas (http://zbbrowser.com) that enables researchers to quickly visualize the larval brain and locate transgenic lines to aid experiments. Such powerful visualization tools facilitate integrated analysis of reconstructed neuronal morphology in the context of the three-dimensional anatomy of the brain (Figure 3).
In order to build a brain atlas, we optimized a protocol that permits very high precision brain registration. We showed that such high precision of alignment permits statistically robust whole-brain analysis of neuronal composition and morphology in zebrafish models of neurological disorders. The technique can be applied to almost any zebrafish neuro-developmental model, thus enabling robust and quantitative detection of subtle changes in brain structure or composition. We recently used the method to test brain structure and composition in zebrafish that carry mutations in genes that are homologous to human genes known to be disrupted in autism-spectrum disorder. By combining analysis of changes in brain structure and startle behavior in these mutants, we can define subsets of autism genes that have similar effects and are therefore likely to act in the same developmental pathways.
Figure 3. Three-dimensional visualization tools facilitate analysis of transgene expression patterns and neuronal morphology in the context of neuroanatomical labels.
Example of a serotonergic neuron from the raphe nucleus (purple) that projects broadly throughout the brain, including into the eminentia granularis (yellow) and closely adjacent to the Mauthner escape neuron (green).
Please use your mouse to interact with this figure. Use the left mouse button to rotate; use the right mouse button or scroll the mouse wheel to zoom; click and hold the mouse wheel to pan.
Additional Funding
- Simons Foundation Autism Research Initiative (2019) #653377 to Dr. Burgess “Generation of zebrafish mutants in homologs of ASD candidate genes”. Concluded.
Publications
- Zoodsma J, Chan k, Golann D, Bhandiwad A, Napoli A, Liu G, Syed S, Burgess HA, Sirotkin H and Wollmuth L. A model to study NMDA receptors in early nervous system development. J Neurosci 2020;40:3631-3645.
- Marquart GD, Tabor KM, Bergeron SA, Briggman KL, Burgess HA. Prepontine non-giant neurons drive flexible escape behavior in zebrafish. PLoS Biol 2019;17:e3000480.
- Horstick EJ, Bayleyen Y, Burgess HA. Molecular and cellular determinants of motor asymmetry in zebrafish. Nat Commun 2020;11:1170.
- Tabor KM, Marquart GD, Hurt C, Smith TS, Geoca AK, Bhandiwad AA, Subedi A, Sinclair JL, Rose HM, Polys NF, Burgess HA. Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping. eLife 2019;8:e42687.
- Luo L, Ambrozkiewizc MC, Benseler F, Chen C, Dumontier E, Falkner S , Furlanis E, Gomez AM, Hoshina N, Huang W, Hutchison MA, Itoh-Maruoka Y, Lavery LA, Li W, Maruo T, Motohashi J, Pai EL, Pelkey KA, Pereira A, Phillips T, Sinclair JL, Stogsdill J, Traunmüller L, Wang J, Wortel J, You W, Abumaria N, Beier KT, Burgess HA, Cepko CL, Cloutier J, Eroglu C, Goebbels S, Kaeser PS, Kay JN, Lu W, Luo L, Mandai K, McBain CJ, Nave K, Prado MA, Prado VF, Rothstein J, Rubenstein JLR, Saher G, Sakimura K, Sanes JR, Scheiffele P, Takai Y, Umemori H, Verhage M, Yuzaki M, Zoghbi HY, Kawabe H, Craig AM. Optimizing nervous system-specific gene targeting with Cre driver lines: prevalence of germline recombination and influencing factors. Neuron 2020;106:37-65.
Collaborators
- Ann M. Craig, PhD, University of British Columbia, Vancouver BC, Canada
- Sinisa Pajevic, PhD, Mathematical and Statistical Computing Lab, CIT, NIH, Bethesda, MD
- Nicholas Polys, PhD, Virginia Polytechnic Institute and State University, Blacksburg, VA
- Howard Sirotkin, PhD, Stony Brook University, Stony Brook, NY
- Lonnie P. Wollmuth, PhD, Stony Brook University, Stony Brook, NY
Contact
For more information, email haroldburgess@mail.nih.gov or visit http://ubn.nichd.nih.gov.