Transcriptional and Translational Regulatory Mechanisms in Nutrient Control of Gene Expression

- Alan G. Hinnebusch, PhD, Head, Section on Nutrient Control of Gene Expression
- Hongfang Qiu, PhD, Staff Scientist
- Jinsheng Dong, PhD, Senior Research Assistant
- Fan Zhang, MS, Senior Research Assistant
- Swati Gaikwad, PhD, Postdoctoral Fellow
- Michelle Gibbs, PhD, Postdoctoral Fellow
- Ritu Gupta, PhD, Postdoctoral Fellow
- Priyanka Mittal, PhD, Postdoctoral Fellow
- Poonam Poonia, PhD, Postdoctoral Fellow
- Priyanka Singh, PhD, Postdoctoral Fellow
- Vishalini Valabhoju, PhD, Postdoctoral Fellow
- Anil Vijjamarri, PhD, Postdoctoral Fellow
- Qiaoyun Zheng, PhD, Postdoctoral Fellow
We study the fundamental mechanisms involved in the assembly and function of translation initiation complexes for protein synthesis, using yeast as a model system in order to exploit its powerful combination of genetics and biochemistry. The translation initiation pathway produces an 80S ribosome bound to mRNA, with methionyl initiator tRNA (tRNAi) base-paired to the AUG start codon. The tRNAi is recruited to the small (40S) subunit in a ternary complex (TC) with the GTP–bound eukaryotic initiation factor eIF2 to produce the 43S preinitiation complex (PIC) in a reaction stimulated by eIFs 1, 1A, 3, and 5. The 43S PIC attaches to the 5′ end of mRNA, facilitated by the cap-binding complex eIF4F (comprising eIF4E, eIF4G, and the RNA helicase eIF4A) and poly(A)–binding protein (PABP) bound to the poly(A) tail, and scans the 5′ untranslated region (UTR) for the AUG start codon. Scanning is promoted by eIF1 and eIF1A, which induce an open conformation of the 40S and rapid TC binding in a conformation suitable for the scanning of successive triplets entering the ribosomal P site (P-out), and by eIF4F and other RNA helicases, such as Ded1 and its paralog Dbp1, that remove secondary structure in the 5′UTR. AUG recognition evokes tighter binding of the TC in the P-in state and irreversible GTP hydrolysis by eIF2, dependent on the GTPase–activating protein (GAP) eIF5, releasing eIF2-GDP from the PIC, with tRNAi remaining in the P site. Joining of the 60S subunit produces the 80S initiation complex ready for protein synthesis. Our current aims in this research area are to: (1) elucidate the functions of eIF1, eIF5, eIF3, and 40S ribosomal proteins in TC recruitment and start-codon recognition; (2) identify distinct functions of the RNA helicases eIF4A (and its cofactors eIF4G/eIF4B), Ded1, and Dbp1, and of the poly(A)–binding protein (PABP) in mRNA activation, 48S PIC assembly, and scanning in vivo; (3) uncover the mechanisms of translational repression and regulation of mRNA abundance by the repressors Scd6, Pat1, the helicase Dhh1, and the mRNA–decapping enzyme Dcp2; (4) elucidate the regulation of Ded1, eIF4G, and Dhh1 functions in response to nutrient limitation or stress; (5) elucidate the in vivo functions of yeast eIF2D orthologs and of the MCT-1/DENR complex in 40S ribosome recycling at stop codons and reinitiation in 3′ untranslated regions in vivo; and (6) elucidate the roles of yeast orthologs of eIF2A and eIF2D in eIF2–independent initiation of translation in stress conditions.
We also analyze the regulation of amino acid–biosynthetic genes in budding yeast as a means of dissecting fundamental mechanisms of transcriptional control of gene expression. During amino acid limitation, transcription of such genes is coordinately induced by the activator Gcn4 as the result of induction of Gcn4 at the translational level. The eviction of nucleosomes that occlude promoter DNA sequences and block access by RNA polymerase is thought to be a rate-limiting step for transcriptional activation. Previous studies implicated certain histone chaperones, ATP–dependent chromatin-remodeling complexes, or histone acetyltransferase (HAT) complexes in eviction of promoter nucleosomes at certain yeast genes, but it is unclear whether these co-factors function at Gcn4 target genes. Our aim is to elucidate the full set of co-factors that participate in promoter nucleosome eviction at Gcn4 target genes, their involvement in this process genome-wide, and the transcriptional consequences of defective nucleosome eviction. Functional cooperation among the chromatin-remodeling complexes SWI/SNF, RSC, and Ino80, as well as the HAT complexes SAGA, NuA4, NuA3, and Rtt109/Asf1, in these processes are under study. We recently discovered that Gcn4 can activate transcription from binding sites within the coding sequences (CDS) of its target genes, inducing internal subgenic sense and antisense (AS) transcripts in addition to the conventional full-length transcripts that initiate 5′ of the CDS; and we are probing both the mechanism and possible regulatory functions of these internal AS transcripts, as well as the roles of co-transcriptional histone methylation, nucleosome reassembly, and mRNA decay enzymes in controlling their synthesis and abundance. We are also probing mechanisms involved in the asymmetric transcriptional induction of genes belonging to pairs of divergently oriented genes where only one gene responds to Gcn4 binding at the shared upstream activation sequences (enhancer).
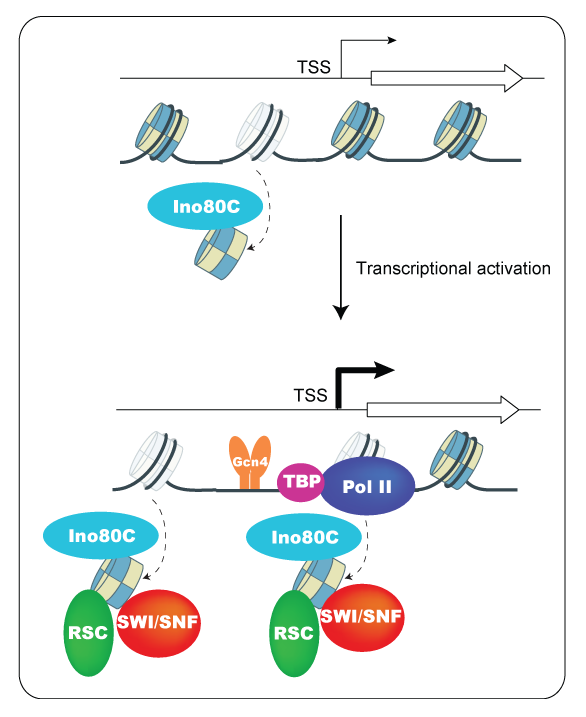
Figure 1. Model for cooperation between the chromatin remodelers Ino80C, SWI/SNF, and RSC in promoter nucleosome disassembly at Gcn4–induced genes
In non-inducing conditions, Ino80C functions in evicting nucleosomes from the promoter region. Induction of Gcn4 leads to additional recruitment of Ino80C, as well as SWI/SNF and RSC, to augment eviction of promoter nucleosomes and thereby increase access of general transcription factors, including TBP, to stimulate PIC assembly by Pol II. Nucleosomes are depicted as multicolored cylinders; cylinders with faint shades indicate nucleosomes undergoing eviction.
eIF2α interactions with mRNA control accurate start-codon selection by the translation preinitiation complex.
Comparison of previous cryo-EM structures of 48S PICs in open, scanning-conducive or closed, arrested conformations revealed interactions unique to the closed complex between Arg residues R55 and R57 of domain 1 of the α-subunit of eIF2 (eIF2α-D1) with mRNA nucleotides 5′ of the AUG codon, including the –3 residue of the “Kozak” context (the Kozak consensus sequence is a nucleic acid motif that functions as the protein translation initiation site in most eukaryotic mRNA transcripts). We showed that substitutions of R55 and R57 reduce recognition of the poor-context AUG codon for SUI1 mRNA (encoding eIF1) and also UUG start codons in Sui– cells (the Ssu– phenotype). We further showed that the R55G-R57E Ssu– substitutions destabilize TC binding to 48S PICs reconstituted with mRNA with a UUG start codon in the in vitro reconstituted system, in the manner expected from specific destabilization of the closed complex at a near-cognate codon. Interestingly, residue R53 of eIF2α-D1 interacts with rRNA residues exclusively in the open complex; we found that the R53E substitution enhances initiation at UUG codons (the Sui– phenotype) and the poor-context SUI1 AUG, and also confers the Gcd– phenotype, indicating constitutively depressed translation of GCN4 mRNA, which results from slow recruitment of the TC to scanning 40S subunits engaged in re-initiation on this mRNA in vivo. In the reconstituted system, R53E stabilized TC binding to UUG complexes while simultaneously reducing the on-rate of TC loading, all in the manner predicted for specific destabilization of the open complex and shift towards the closed state. We conclude that distinct interactions of eIF2α-D1 with the rRNA or mRNA stabilize first the open, and then the closed, conformation of the PIC to regulate the accuracy and efficiency of start codon selection in vivo [Reference 5].
eIF1 discriminates against suboptimal initiation sites to prevent excessive uORF translation genome-wide.
To uncover the genome-wide role of eIF1 in promoting accurate start-codon selection, we conducted ribosome profiling (deep-sequencing of all native ribosome-protected mRNA fragments) of the eIF1–L96P mutant, which is impaired for interactions with eIF3c, eIF5, and the 40S subunit, expected to occur in the scanning PIC. The profiling data indicated that L96P increases initiation at near-cognate start codons (NCCs) that initiate N-terminal extensions for several proteins, which we confirmed by reporter assays. L96P also increased translation of hundreds of upstream open reading frames (uORFs) initiated by NCCs. We also observed increased utilization of poor-context AUGs, both at AUG–initiated uORFs (that exhibit poor context as a group) and at the small fraction of main CDSs with poor-context AUGs. Interestingly, L96P leads to reduced translation of a subset of mRNAs in a manner associated with increased uORF translation, including carboxypeptidase A1 (CPA1) mRNA, which is negatively regulated by its single AUG uORF. Thus, eIF1 acts broadly to discriminate against NCC start codons and poor-context AUGs; impairing this function can increase the repressive effects of uORFs and alter the ratios of protein isoforms translated from the same mRNAs by NCC versus AUG start codons [Reference 1].
Functional interplay between RNA helicases Ded1 and Dbp1 in stimulating translation of structured mRNAs in vivo
RNA helicases eIF4A and Ded1 are believed to resolve mRNA structures that impede ribosome attachment or scanning to the start codon. By ribosome profiling of mutations in Ded1 or eIF4A, we previously found that inactivation of Ded1 reduced the relative translational efficiencies (TEs) of over 600 mRNAs characterized by relatively long and structured 5′UTRs, whereas inactivation of eIF4A similarly affected only about 40 mRNAs. Thus, Ded1 is critically required for PIC attachment and scanning through secondary structures, while eIF4A promotes a step of initiation common to all mRNAs. We reconstituted the function of Ded1 in a purified system by showing that Ded1 accelerates 48S PIC assembly to a greater extent for Ded1–hyperdependent versus Ded1–hypodependent mRNAs identified by ribosome profiling, and that eliminating 5′UTR stem-loop structures enhanced Ded1–independent recruitment and diminished Ded1 acceleration of 48S assembly. To illuminate the in vivo function of the Ded1 paralog Dbp1, we conducted ribosome profiling of dbp1Δ and dbp1Δded1 double mutants. The results indicate that Dbp1 functionally cooperates with Ded1 throughout the translatome in stimulating translation of mRNAs with long, structure-prone 5′UTRs, as the TE reductions in the double mutant generally exceed those in the ded1 single mutant. For many such mRNAs, Dbp1 largely masks the involvement of Ded1. Importantly, Dbp1 mimics Ded1 in accelerating 48S preinitiation complex (PIC) assembly in the reconstituted system on Ded1–hyperdependent mRNAs with structured 5′UTRs. Using the recently developed method of TCP-seq for genome-wide profiling of 40S subunits, we quantified PIC occupancies in 5′UTRs and found that 40S subunits tend to accumulate in the 5′UTRs of mRNAs in the helicase mutants, particularly for mRNAs judged to be Ded1/Dbp1–hyperdependent by 80S ribosome profiling, thus providing direct evidence that Ded1/Dbp1 stimulate scanning through structured 5′UTRs in vivo to enhance translation. We also uncovered cooperation between these helicases in promoting 43S PIC attachment to a subset of helicase-dependent mRNAs, which exhibit reduced 40S occupancies in 5′UTRs in the helicase double mutant [Reference 6].
eIF4A and eIF4E interactions with distinct residues of the Ded1 N-terminus stimulate Ded1 function in translation initiation in vivo.
Binding of eIF4F to the mRNA cap structure enhances recruitment of the 43S PIC to the 5′end and subsequent scanning of the 5′UTR. Ded1 physically interacts with eIF4A and the eIF4G subunit of eIF4F, and eIF4A and eIF4G can both stimulate unwinding of a model RNA substrate by Ded1 in vitro. Previously, we showed that the Ded1 C-terminal domain (CTD) and its two interacting domains in eIF4G, dubbed RNA2 and RNA3, and the Ded1 N-terminal domain (NTD) that interacts with eIF4A, all enhance Ded1 stimulation of 48S PIC assembly in the reconstituted in vitro system. Ded1 also interacts with eIF4E; however the binding sites for eIF4A and eIF4E in the Ded1–NTD were unknown. By substituting blocks of conserved residues in the Ded1–NTD, we found that alanine replacements of residues 21–27 and 51–57 reduce Ded1 binding to eIF4A in vitro, impair association between native Ded1 and eIF4A in cell extracts, reduce cell growth, bulk translation initiation, and translation of Ded1–hyperdependent reporter mRNAs harboring stem-loop insertions. Overexpressing eIF4A diminished the growth defects for each single substitution, but not for the 21–27/51–57 double substitution, which is null for eIF4A binding, supporting the importance of Ded1–NTD/eIF4A interaction in cells. Substituting the non-overlapping residues 59–65 and 83–89 reduced Ded1–NTD binding to eIF4E in vitro, as well as Ded1–eIF4E association in extracts, and conferred reduced translation of the Ded1–hyperdependent reporters. Combining all four NTD substitutions conferred an additive growth defect indistinguishable from deletion of the NTD, suggesting that eIF4A/eIF4E binding is the key in vivo function of the Ded1 NTD. Deleting the Ded1–CTD impairs growth only when combined with NTD substitutions, implying that the Ded1–CTD interaction with eIF4G is dispensable when Ded1 can interact with eIF4A and eIF4E. In the reconstituted system, the Ded1 NTD substitutions that eliminate eIF4A binding reduce the maximal rate of 48S PIC assembly on a Ded1–dependent mRNA harboring a 5′UTR SL, and also increase the amount of Ded1 required to achieve the half-maximal rate (K1/2). Disruption of the Ded1–NTD/eIF4E interaction has a similar effect of elevating the Ded1 K1/2 for 48S assembly. The findings support the notion that Ded1 NTD interactions with eIF4A and eIF4E stabilize a Ded1–eIF4E–eIF4G–eIF4A quaternary complex that enhances Ded1’s ability to resolve secondary structures in 5′UTRs [Reference 2].
Chromatin remodeler (CR) Ino80C acts independently of histone variant H2A.Z to evict promoter nucleosomes and stimulate transcription of highly expressed genes.
The CR Ino80C was found to be sufficient for reconstituting a near-native nucleosomal organization in vitro, and was also implicated in nucleosome editing to replace the histone variant H2A.Z (encoded by HTZ1) with canonical H2A. The removal of an H2A.Z:H2B dimer by Ino80C could render the partially disassembled nucleosome more susceptible to eviction; however, a prominent role for Ino80C in promoter nucleosome eviction had not been reported. By ChIP-seq analysis of the histone H3 and RNA polymerase II (Pol II) in an ino80Δ mutant lacking the Ino80C catalytic subunit, we found that Ino80C functions on par with SWI/SNF in eviction of promoter nucleosomes and transcriptional activation of Gcn4 target genes, and plays a much greater role than the chromatin-remodeling complexes SWI/SNF at a group of several hundred Ino80C–hyperdependent genes. Compared with both SWI/SNF and RSC, Ino80C generally functions over a wider interval spanning the -1 and +1 nucleosomes, the nucleosome-depleted-region (NDR), and NDR–proximal genic nucleosomes. At Gcn4 target genes, the degree of nucleosome eviction defect is correlated with the reduction in transcription; and ChIP-seq analysis of the GTF TATA–binding protein (TBP) revealed that defects in nucleosome eviction are accompanied by reduced promoter occupancies of TBP, and hence PIC assembly. ChIP-seq analysis of Ino80 itself shows that Ino80C is enriched at both Gcn4 target genes and Ino80C–hyperdependent genes. Thus, Ino80C cooperates with RSC and SWI/SNF in evicting promoter nucleosomes to enhance PIC assembly and transcription at many highly expressed genes [Reference 4] (see image below).
If Ino80C enhances nucleosome eviction strictly in the course of editing H2A.Z-H2B dimers, then deleting HTZ1 should mimic the effect of deleting Ino80 on promoter nucleosome eviction. Moreover, depleting Ino80 should have no effect on nucleosome occupancies in cells lacking HTZ1. At odds with these predictions, we found that the htz1Δ mutation has much smaller effects than ino80Δ on eviction of promoter nucleosomes. Moreover, depleting Ino80 from the nucleus by “anchor-away” impaired histone eviction in cells lacking HTZ1. Thus, Ino80C can function like the SWI/SNF family members SWI/SNF and RSC in promoting chromatin access independently of nucleosome editing [Reference 4].
Function and regulation of Gcn4 binding within coding regions in activating internal and canonical 5′ promoters in yeast
We are also interested in determining the role of promoter nucleosome eviction in controlling binding of Gcn4 itself upstream from the promoters of its target genes. We thus set out to define all binding sites for Gcn4 throughout the genome in wild-type (WT) cells. ChIP-seq analysis of Gcn4 binding revealed 546 genomic sites occupied by Gcn4 in starved cells, representing only 30% of all genomic sequences with significant matches to the consensus Gcn4–binding motif. Analysis of nucleosome occupancy data from MNase-seq analysis revealed that the distance of a motif from the nearest nucleosome dyad and its match to the consensus sequence are the major determinants of Gcn4 binding in vivo. Surprisingly, only 40% of the bound sites are in promoters, and analysis of genome-wide mRNA expression data and ChIP-seq analysis of Pol II in starvation conditions indicates that only 60% of such promoter-located Gcn4 occupancy peaks activate transcription, indicating extensive negative control over Gcn4 function. Remarkably, most of the remaining 300 Gcn4–bound motifs reside within coding sequences (CDS), with 75 representing the only bound motif in the vicinity of a known Gcn4–induced gene. RNA-seq analysis revealed that many such unconventional Gcn4 occupancy peaks map between divergent antisense (AS) and sub-genic sense transcripts induced from within CDS under starvation conditions, and are also located adjacent to starvation-induced TBP occupancy peaks detected by ChIP-seq analysis, findings that are consistent with Gcn4 activation of cryptic, bidirectional internal promoters at these genes. Mutational analysis confirmed that Gcn4–bound motifs within CDS can activate both sub-genic and full-length transcripts from the same or adjacent genes, demonstrating that functional Gcn4 binding is not confined to promoters. Our results show that internal promoters can be regulated by a well-defined activator that also functions at conventional 5′-positioned promoters.
Current experiments are aimed at determining whether induction of internal AS promoters by Gcn4 serves to dampen its activation of the upstream promoters for the full-length sense transcripts at these genes, via collisions between Pol II molecules transcribing from opposite DNA strands, or by co-transcriptional methylation by the histone-lysine N-methyltransferases Set1 or Set2, with attendant deacetylation of the upstream promoter nucleosomes. We are also examining the roles of Set1 and Set2, and the histone chaperone Spt6 in suppressing cryptic internal promoters activated by Gcn4, and of the nuclear exosome and the nonsense-mediated decay (NMD) pathway in degradation of the AS transcripts. We are also probing the roles of chromatin remodelers, HAT complexes, and general regulatory factors in promoting Gcn4 binding to its target sequences in vivo.
Publications
- Zhou F, Zhang H, Kulkarni S, Lorsch JR, Hinnebusch AG. eIF1 discriminates against suboptimal initiation sites to prevent excessive uORF translation genome-wide. RNA 2020;26:419-438.
- Gulay S, Gupta N, Lorsch JR, Hinnebusch AG. 225. Distinct interactions of eIF4A and eIF4E with RNA helicase Ded1 stimulate translation in vivo. eLife 2020;e58243.
- Wagner S, Herrmannová A, Hronová V, Gunišová S, Sen ND, Hannan RD, Hinnebusch AG, Shirokikh NE, Preiss T, Valášek LS. Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol Cell 2020;79:546-560.
- Qiu H, Biernat E, Govind CK, Rawal Y, Chereji RV, Clark DJ, Hinnebusch AG. Chromatin remodeler Ino80C acts independently of H2A.Z to evict promoter nucleosomes and stimulate transcription of highly expressed genes in yeast. Nucleic Acids Res 2020;48:8408-8430.
- Thakur A, Gaikwad S, Vijjamarri AK, Hinnebusch AG. eIF2a interactions with mRNA control accurate start codon selection by the translation preinitiation complex. Nucleic Acids Res 2020;48:10280-10296.
- Sen ND, Gupta N, K Archer S, Preiss T, Lorsch JR, Hinnebusch AG. Functional interplay between DEAD-box RNA helicases Ded1 and Dbp1 in preinitiation complex attachment and scanning on structured mRNAs in vivo. Nucleic Acids Res 2019;47(16):8785-8806.
Collaborators
- Stuart Archer, PhD, Monash Bioinformatics Platform, Monash University, Australia
- David Clark, PhD, Section on Chromatin and Gene Expression, NICHD, Bethesda, MD
- Chhabi Govind, PhD, Oakland University, Rochester, MI
- Jon Lorsch, PhD, Laboratory on the Mechanism and Regulation of Protein Synthesis, NICHD, Bethesda, MD
- Thomas Preiss, PhD, The John Curtin School of Medical Research, The Australian National University, Canberra, Australia
- Venkatraman Ramakrishnan, PhD, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom
Contact
For more information, email hinnebua@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/hinnebusch.