Mechanisms of Disease in Preterm Labor and Complications of Prematurity; Prenatal Diagnosis of Congenital Anomalies

- Roberto Romero, MD, DMedSci, Chief, Perinatology Research Branch
Preterm birth is the leading cause of perinatal morbidity and mortality worldwide. The cost of prematurity in the U.S. alone is estimated to be $26 billion per year. An important goal is to understand the mechanisms of disease responsible for spontaneous preterm birth and fetal injury and to develop methods for the prediction and prevention of preterm birth.
The Perinatology Research Branch (PRB) proposed that preterm parturition is a syndrome caused by many pathologic processes, i.e., that preterm labor is one syndrome but has many causes. The emphasis of our Branch is to study intra-amniotic infection and inflammation, vascular disorders, maternal anti-fetal rejection (chronic inflammatory lesions of the placenta), cervical disease, and a decline in progesterone action. Previously, we reported that intra-amniotic inflammation, which affects at least one out of every three preterm neonates, is characterized by the activation of amniotic-fluid neutrophils, cells that represent the first line of defense against infection. Using DNA fingerprinting, we determined that amniotic-fluid neutrophils are of fetal origin in cases of preterm labor, maternal origin in cases of clinical chorioamnionitis at term, and mixed origin in patients who have inflammatory processes near term. Moreover, in a series of studies, we were able to demonstrate that neutrophils produce anti-microbial peptides and exhibit the formation of extracellular traps, whereby they immobilize and kill bacteria.
The Branch also studies other obstetrical syndromes that account for the high rate of infant mortality in the United States, including clinical chorioamnionitis, which is the most common infection-related diagnosis in delivery units around the world, as well as meconium aspiration syndrome and amniotic fluid embolism.
Congenital anomalies continue to be a leading cause of perinatal mortality in the U.S. Imaging, a powerful tool for scientific discovery, has changed the practice of obstetrics and maternal-fetal medicine. Imaging with ultrasound permits the definition of fetal anatomy, biometry, growth, and the study of physiologic parameters, such as cardiac function, fetal sleep, and breathing. We reported that fetal intelligent navigation echocardiography (FINE) can be used to screen for congenital heart disease and we addressed the frequency with which a dataset can be obtained in the second and third trimester of pregnancy. We were able to obtain an informative dataset in approximately 72% of cases, which indicates that the method can be used in an unselected population. Moreover, we reported the prenatal diagnosis of dextrocardia using FINE. The technology has been licensed and is now commercially available to sonographers worldwide.
Regulatory T cells and adverse neonatal outcomes
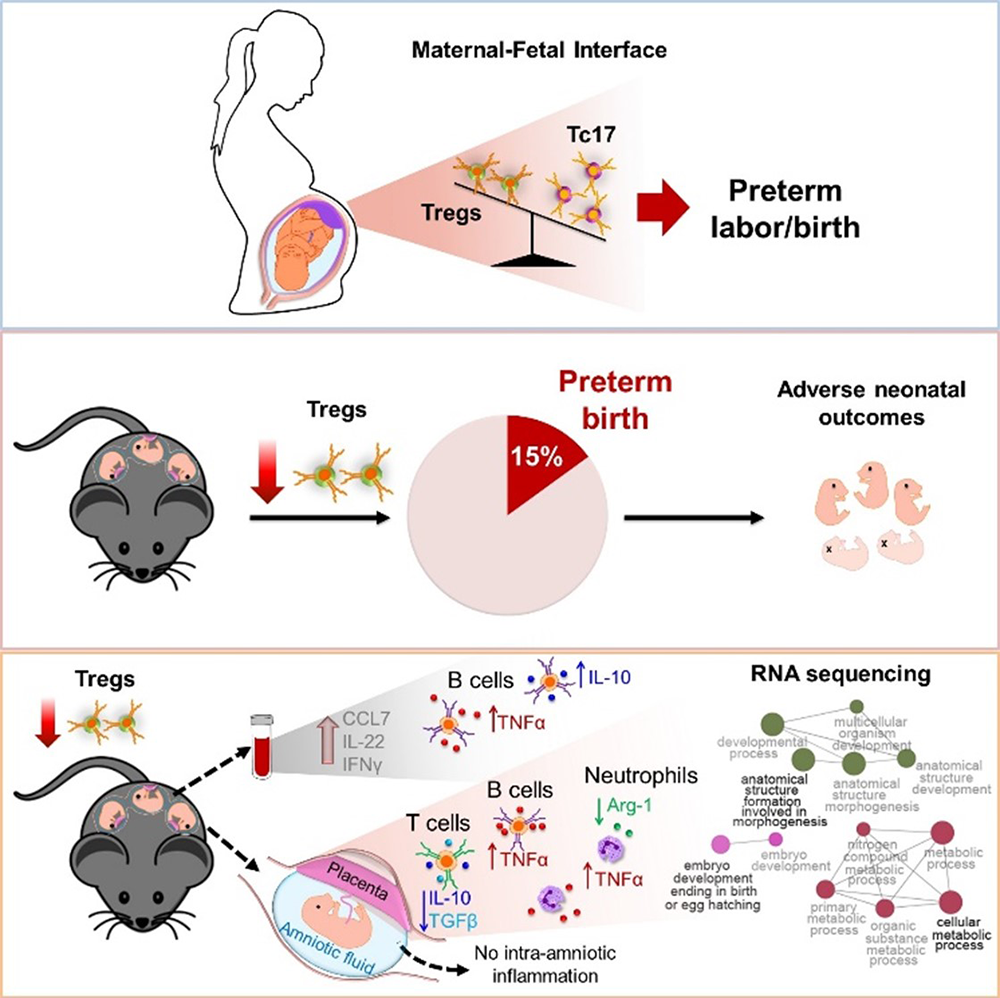
Regulatory T cells emerge in placental mammals to enforce maternal-fetal tolerance; therefore, most research has focused on understanding their role during early pregnancy. However, their role in the third period of gestation had not been mechanistically investigated. We undertook an extensive investigation, which included both human and animal models, to identify a role for regulatory T cells in the pathophysiology of preterm labor/birth and adverse neonatal outcomes. First, we showed that women with idiopathic preterm labor/birth had reduced proportions of functional regulatory T cells at the maternal-fetal interface. Next, we showed that the depletion of regulatory T cells led to preterm birth in a subset of cases. More importantly, the depletion led to growth restriction in both preterm and term neonates, deleterious effects that were ameliorated by the restoration of such cells to the mother. The immune mechanisms implicated in adverse perinatal outcomes induced by the depletion of regulatory T cells involved: first, a mild systemic inflammatory response mediated by CCL7 and IL-22; second, specific innate and adaptive immune responses in the decidua, myometrium, and placenta; and third, dysregulation of developmental and cellular processes in the placenta; these all occurred in the absence of intra-amniotic inflammation. The study represents the first mechanistic evidence supporting a role for regulatory T cells in spontaneous preterm labor and birth as well as impaired growth of the offspring (Figure 1).
Figure 1.
Regulatory T cells play a central role in preterm labor and birth and in adverse neonatal outcomes.
Prevention of preterm birth and neonatal mortality with clarithromycin
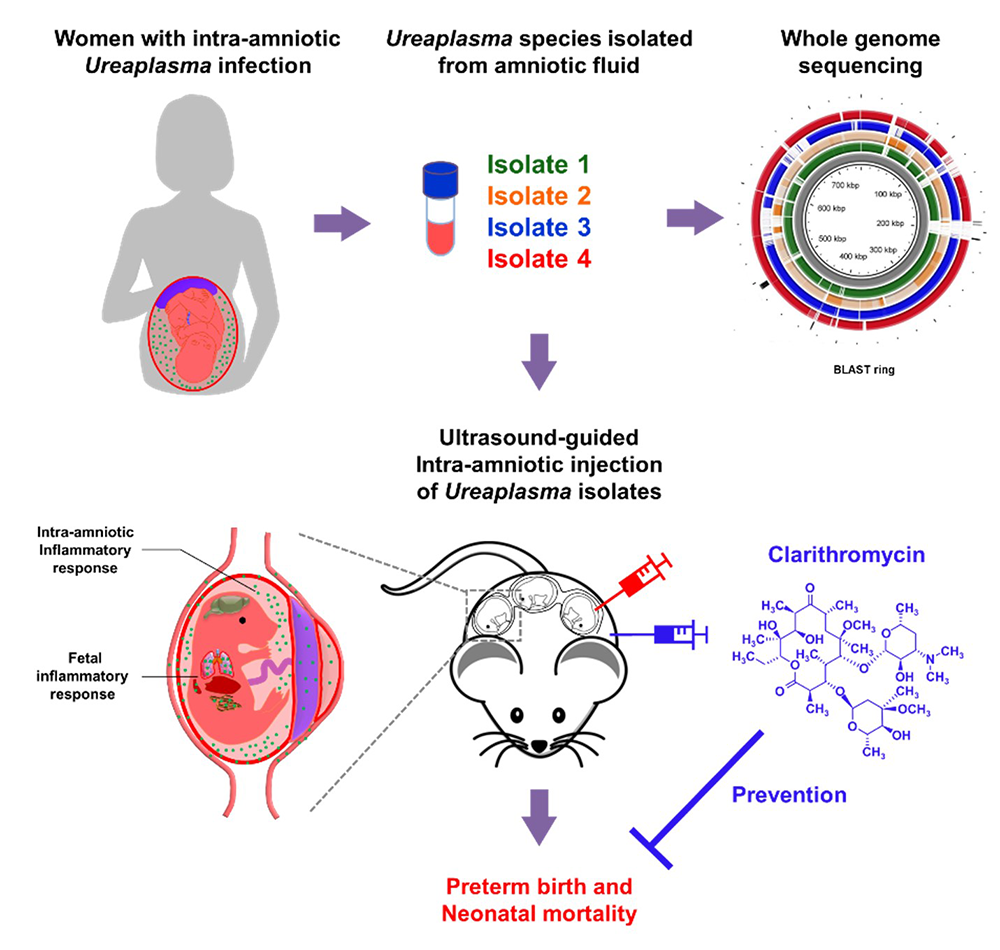
One of every four preterm neonates is born to women with intra-amniotic infection, a clinical condition commonly associated with invasion of the amniotic cavity by Ureaplasma species. Yet, little is known about the taxonomy of and host immune response against these bacteria. We applied a multifaceted approach, including human amniotic fluid samples, in vivo models, and in vitro manipulations, to study the maternal-fetal immunobiology of Ureaplasma infection during pregnancy and a strategy to treat this condition. First, we performed a taxonomic characterization of Ureaplasma isolates from women with intra-amniotic infection, which revealed that Ureaplasma parvum is the most common bacterium found in this clinical condition. Next, we showed, in using animal models, that the intra-amniotic inoculation of Ureaplasma isolates induced varying rates of preterm birth and, more importantly, induced high rates of mortality in preterm and term neonates. Regardless of their potency to induce preterm birth, Ureaplasma isolates were capable of inducing a severe inflammatory response in the amniotic cavity and fetus. Notably, treatment with clarithromycin, a recently recommended yet not widely utilized antibiotic, prevented preterm birth and neonatal mortality induced by the intra-amniotic inoculation of Ureaplasma parvum. The investigations give insight into the maternal-fetal immunobiology of intra-amniotic infection and provide a critical demonstration of the effectiveness of clarithromycin to treat this clinical condition (Figure 2).
Figure 2.
Intra-amniotic infection with Ureaplasma parvum causes preterm birth and neonatal mortality that are prevented by treatment with clarithromycin.
The efficacy of cervical pessary to prevent preterm birth
We performed a meta-analysis to assess the efficacy and safety of cervical pessary to prevent preterm birth and adverse perinatal outcomes in asymptomatic high-risk women. A total of 12 studies involving 4687 women and 7,167 fetuses/infants were included in the review. Overall, there were no significant differences between the pessary and no-pessary groups in the risk of spontaneous preterm birth at less than 34 weeks of gestation among singleton gestations with a short cervix (cervical length of 25 mm or less), unselected twin gestations, twin gestations with a cervical length of less than 38 mm, and twin gestations with a cervical length of 25 mm or less. Overall, no significant differences were observed between the pessary and no-pessary groups in preterm birth at less than 37, 32, or 28 weeks of gestation, and most adverse pregnancy, maternal, and perinatal outcomes. Vaginal discharge was significantly more frequent in the pessary group than in the no-pessary group. In conclusion, current evidence does not support the use of cervical pessary to prevent preterm birth or to improve perinatal outcomes in singleton or twin gestations with a short cervix and in unselected twin gestations.
Single-cell transcriptional signatures of the human placenta
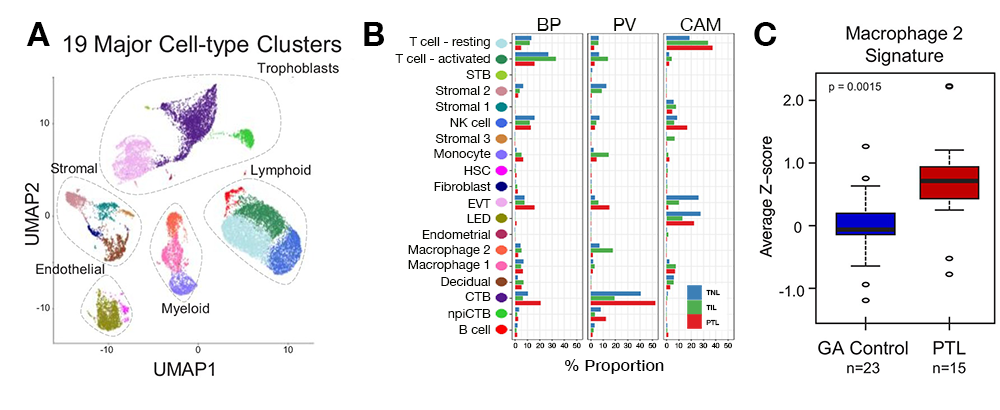
Understanding human parturition is essential to tackle the challenge of prematurity, which affects 15 million neonates every year. We investigated the human placenta at single-cell resolution in spontaneous labor at term and in preterm labor, having completed the first molecular characterization of the common pathway of preterm and term parturition. The key observations are as follows. First, we characterized, for the first time, the common molecular pathway of human parturition (spontaneous term labor and preterm labor) at single-cell resolution, where most of the differentially expressed genes were found in maternal macrophages from the chorioamniotic membranes and were involved in immune and inflammation-related pathways (Figure 3). Second, in the context of preterm labor, we found that most differentially expressed genes were detected in trophoblast cell types. Third, we identified two new cell types: maternal lymphatic endothelial decidual cells in the chorioamniotic membranes; and non-proliferative interstitial cytotrophoblasts in the placental villi (Figures 3A and 3B). Fourth, we identified placental scRNA-Seq transcriptional signatures in the maternal circulation, revealing that the cellular dynamics of the placenta can be reflected in maternal blood total RNA and, therefore, can be monitored throughout pregnancy (Figure 3C).
Figure 3. Transcriptional map of the placenta in human parturition
A. Uniform Manifold Approximation Plot (UMAP), where dots represent single cells and colors represent cell type.
B. Average proportions of cell types by placental compartments (BP: basal plate; PV: placental villi; CAM: chorioamniotic membranes) and study groups (TNL: term no labor; TIL: term in labor; PTL: preterm labor). STB: syncytiotrophoblast; EVT: extravillous trophoblast; CTB: cytotrophoblast; HSC: hematopoietic stem cell; npiCTB: non-proliferative interstitial cytotrophoblast; LED: lymphoid endothelial decidual cell.
C. Quantification of scRNA-Seq signatures in maternal circulation. Perturbations in scRNA-seq macrophage signature in women with spontaneous PTL compared with gestational age-matched (GA) controls.
The human placenta and SARS-CoV-2
Using single-cell gene expression, we determined that co-expression of the ACE2 and TMPRSS2 transcripts is negligible in the placenta and chorioamniotic membranes. ACE2 and TMPRSS2 are the major proteins used by SARS-CoV-2 to infect the cells, spread, and cause COVID-19 disease; yet, they are rarely co-transcribed in any of the cell-types of the placenta or at any of the time points spanning the three trimesters of pregnancy. In contrast, the transcripts for receptors of other viruses known to cause fetal infection, such as Zika and cytomegalovirus, were found in larger quantities in placental cells. The finding helps to explain why SARS-CoV-2 is rarely vertically transmitted from mother to fetus.
The cellular transcriptome in the maternal circulation during pregnancy
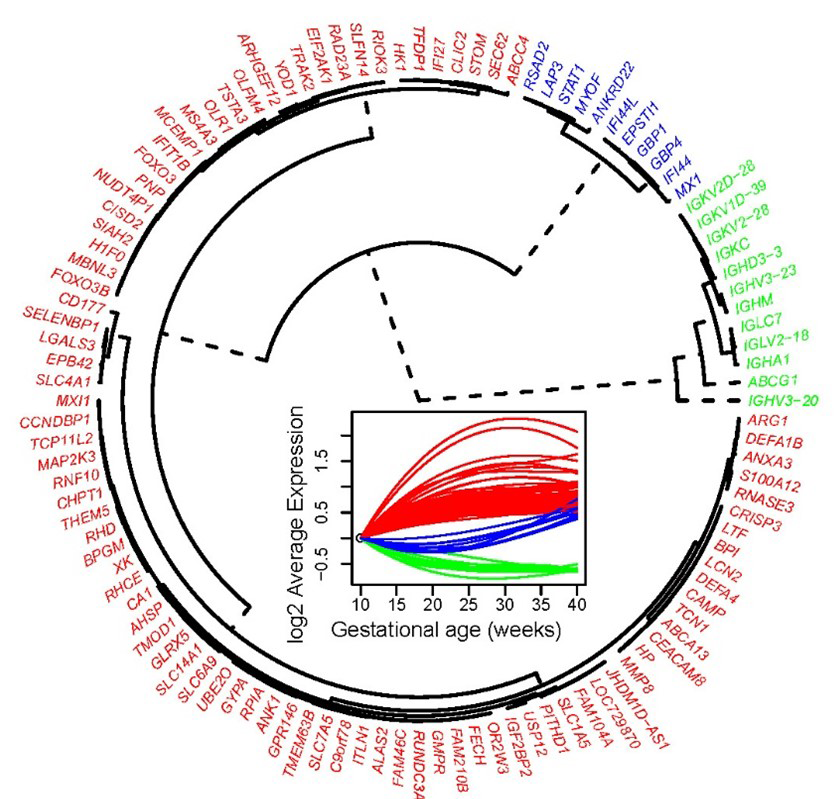
Pregnancy represents a unique immunological state, in which the mother adapts to tolerate the semi-allogenic conceptus; yet, the cellular dynamics in the maternal circulation are poorly understood. Using exon-level expression profiling of up to six longitudinal whole blood samples from 49 pregnant women we found that: (1) the strongest expression changes followed three distinct longitudinal patterns, with genes related to host immune response (e.g., MMP8, DEFA1B, DEFA4, LTF) showing a steady increase in expression from 10 to 40 weeks of gestation (see red cluster in Figure 4); (2) many biological processes and pathways related to immunity and inflammation were modulated during gestation; (3) the expression of mRNA signatures of T cells, B cells, and erythroid cells, defined using single-cell genomics, followed unique patterns during gestation; and (4) significant whole-blood mRNA and plasma protein correlations were observed for genes that are part of the T cell signature. The findings provide insight into the immunobiology of normal pregnancy and suggest candidate predictors for dating gestational age.
Figure 4.
Whole-blood gene-expression longitudinal-trajectory clusters for genes most highly modulated throughout gestation in normal pregnancy
New and advanced features of fetal intelligent navigation echocardiography (FINE)
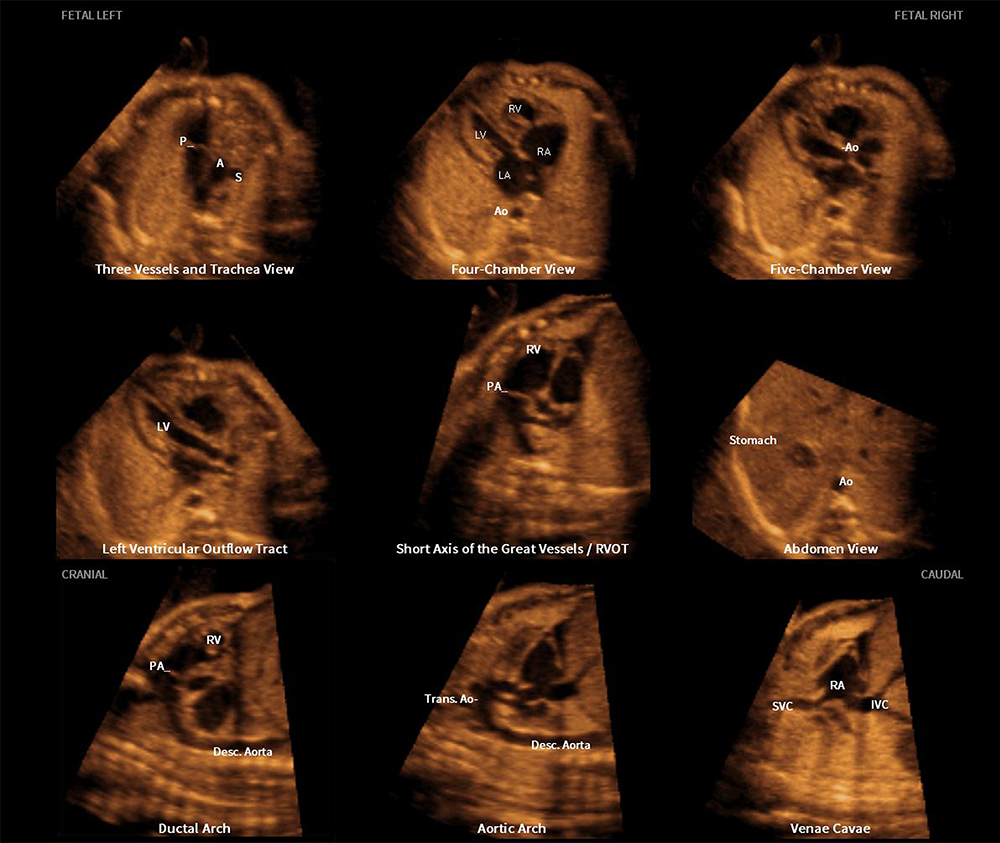
Congenital heart disease (CHD) is the leading organ-specific birth defect, as well as the leading cause of infant morbidity and mortality from congenital malformations. Therefore, a comprehensive screening examination of the fetal heart should be performed in all women to maximize the detection of CHD. Four-dimensional sonography with spatiotemporal image correlation (STIC) technology displays a cine loop of a complete single cardiac cycle in motion. A novel method known as fetal intelligent navigation echocardiography (FINE) was previously developed to interrogate STIC volume datasets using intelligent navigation technology. The method permits the automatic display of nine standard fetal echocardiography views, required to diagnose most cardiac defects. FINE considerably simplifies fetal cardiac examinations and reduces operator dependency. It has both high sensitivity and specificity for the detection of CHD. Indeed, FINE has been integrated into several commercially available ultrasound platforms. Recently, eight novel and advanced features were developed for the FINE method. Such features can be categorized based upon their broad goals. The first goal is to simplify FINE further, and consists of the following features: (1) auto fetal positioning (or FINE align); (2) skip points (to allow quantitative measurements to be performed on the cardiac views generated by FINE); (3) predictive cursor; (4) static mode volume (Figure 5); (5) breech sweep; (6) automatic cardiac axis; and (7) cardiac biometry. The last goal is to improve the success of obtaining fetal echocardiography view(s) and consists of (8) Maestro planar navigation.
Figure 5. Application of the FINE method to a fetus with a normal heart (Static mode volume)
Three-dimensional static volume obtained in a very rapid acquisition time (1 second) and characterized by a high frame rate, which leads to a dataset with improved quality. Nine normal cardiac diagnostic planes in a single template are depicted by FINE. Note the unique feature of automatic labeling (through intelligent navigation) of each plane, anatomic structures, fetal left and right sides, and cranial and caudal ends.
Optical ultrasound simulation–based training in obstetric sonography
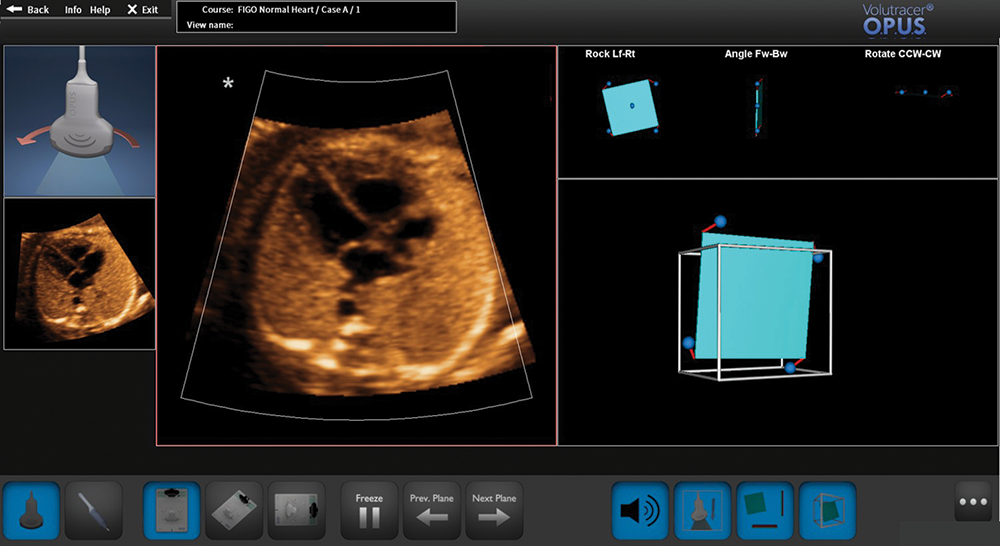
Ultrasound is an imaging modality that is highly operator dependent. This year, we published a review of the challenges in learning how to perform obstetric sonography and the processes necessary to acquire expert performance skills in sonography; we evaluated simulation-based education and learning, and the value of medical simulation. Ultrasound simulators are an effective means of teaching obstetric sonography, because it provides training, deliberate practice, and performance evaluation/feedback, which allows continuous and critical self-evaluation. We reviewed evidence that simulation can improve performance in obstetric ultrasound examination, reviewed current simulators, and discussed the current problems/gaps in ultrasound simulation. Optical positioning ultrasound simulation is a novel high-fidelity simulation learning system, which addresses many of these problems/gaps, and we introduced this for the first time in this article. The technology uses a camera-based optical tracking system and has a limited amount of hardware, making the simulator compact, portable, and personal. A computer or laptop is used to load actual (vs. artificially created) ultrasound cases, which are organ-specific. The goal of the trainee is to achieve a sonographic target plane. The Guided mode (Figure 6) uniquely provides simulator voice commands that instruct the user on how to manipulate the transducer, contextual videos and animation, and graphic feedback to guide the user towards obtaining the correct target plane of interest (e.g., four-chamber view).
Figure 6. Scanning the fetal heart through the Guided mode of the optical positioning ultrasound simulator
A four-dimensional spatiotemporal image correlation volume dataset has been loaded onto the computer. The computer screen depicts the following: (1) target plane (small image of four-chamber view) located on the left side of the screen; (2) plane being generated by the virtual transducer (large image of the five-chamber view); (3) visual graphic (left upper corner) provides instructions via contextual video and animation on how to move the virtual transducer to achieve the target plane; (4) inclinometer (upper right panel showing three diagrams); and (5) cube (lower right panel).
The amniotic fluid cell-free transcriptome
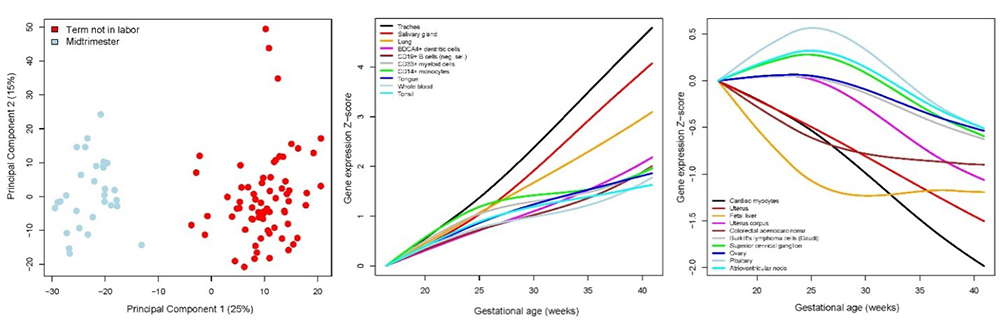
The amniotic fluid (AF) cell-free RNA (cfRNA) is modulated with physiologic and pathologic processes during pregnancy. We therefore sought to define AF normal expression and splicing patterns to aid in identifying biomarkers for assessing fetal development and obstetrical disease. About one hundred AF transcriptomes were profiled in samples collected during midtrimester and at term, and we quantified the expression of tissue-specific and cell-type–specific signatures defined by single-cell genomics. We found that 11% of the coding and 6% of the non-coding AF genes were differentially expressed between midtrimester and term gestation. Expression changes with advancing gestation featured increased expression of genes specific to the trachea, salivary glands, and lung; and decreased expression of genes specific to the cardiac myocytes, uterus, and fetal liver, among others. We also found that, with advancing gestation, differential splicing involved genes related to brain development and immunity pathways, including some that were missed based on differential expression analysis alone. This is thus the largest AF transcriptomics study in normal pregnancy, reporting for the first time that single-cell genomic signatures can be tracked in the AF and display complex patterns of expression during gestation. We also demonstrated a role for alternative splicing in tissue-identity acquisition, organ development, and immune processes. The results may have implications for the development of fetal testing to assess placental function and fetal organ maturity (Figure 7).
Figure 7. Amniotic fluid cell-free transcriptome in normal pregnancy
Principal components analysis (left); tissues and cell-type signatures with increasing (center); and decreasing (right) expression patterns with advancing gestational age.
Publications
- Gomez-Lopez N, Arenas-Hernandez M, Romero R, Miller D, Garcia-Flores V, Leng Y, Xu Y, Galaz J, Hassan SS, Hsu CD, Tse H, Sanchez-Torres C, Done B, Tarca AL. Regulatory T cells play a role in a subset of idiopathic preterm labor/birth and adverse neonatal outcomes. Cell Rep 2020;32:107874.
- Motomura K, Romero R, Xu Y, Theis KR, Galaz J, Winters AD, Slutsky R, Garcia-Flores V, Zou C, Levenson D, Para R, Ahmad MM, Miller D, Hsu CD, Gomez-Lopez N. Intra-amniotic infection with ureaplasma parvum causes preterm birth and neonatal mortality that are prevented by treatment with clarithromycin. mBio 2020;11:e00797-20.
- Conde-Agudelo A, Romero R, Nicolaides KH. Cervical pessary to prevent preterm birth in asymptomatic high-risk women: a systematic review and meta-analysis. Am J Obstet Gynecol 2020;223:42-65.e2.
- Pique-Regi R, Romero R, Tarca AL, Luca F, Xu Y, Alazizi A, Leng Y, Hsu CD, Gomez-Lopez N. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? eLife 2020;9:e58716.
- Gomez-Lopez N, Romero R, Hassan SS, Bhatti G, Berry SM, Kusanovic JP, Pacora P, Tarca AL. The cellular transcriptome in the maternal circulation during normal pregnancy: a longitudinal study. Front Immunol 2019;10:2863.
- Tarca AL, Romero R, Pique-Regi R, Pacora P, Done B, Kacerovsky M, Bhatti G, Jaiman S, Hassan SS, Hsu CD, Gomez-Lopez N. Amniotic fluid cell-free transcriptome: a glimpse into fetal development and placental cellular dynamics during normal pregnancy. BMC Med Genomics 2020;13:25.
Collaborators
- Tinnakorn Chaiworapongsa, MD, Wayne State University School of Medicine, Detroit, MI
- Agustin Conde-Agudelo, MD, Wayne State University School of Medicine, Detroit, MI
- Mark Haacke, PhD, Wayne State University School of Medicine, Detroit, MI
- Leonid Margolis, PhD, Section on Intercellular Interactions, NICHD, Bethesda, MD
- Adi L. Tarca, PhD, Wayne State University, Detroit Medical Center, Detroit, MI
- Lami Yeo, MD, Wayne State University School of Medicine, Detroit, MI
- Bo Hyun Yoon, MD, PhD, Seoul National University, Seoul, Korea
Contact
For more information, email romeror@mail.nih.gov or visit http://irp.nih.gov/pi/roberto-romero.