The Molecular Mechanics of Eukaryotic Translation Initiation

- Jon Lorsch, PhD, Chief, Laboratory on the Mechanism and Regulation of Protein Synthesis
- Jagpreet Nanda, PhD, Staff Scientist
- Fujun Zhou, PhD, Research Fellow
- Shardul Kulkarni, PhD, Postdoctoral Intramural Research Training Award Fellow
The goal of our research is to elucidate the molecular mechanisms underlying the initiation phase of protein synthesis in eukaryotic organisms. We use the yeast Saccharomyces cerevisiae as a model system and employ a range of approaches, from genetics to biochemistry to structural biology, in collaboration with Alan Hinnebusch's and Tom Dever’s labs and several other research groups around the world.
Eukaryotic translation initiation is a key control point in the regulation of gene expression. It begins when an initiator methionyl tRNA (Met-tRNAi) is loaded onto the small (40S) ribosomal subunit. Met-tRNAi binds to the 40S subunit as a ternary complex (TC) with the GTP–bound form of the initiation factor eIF2. Three other factors, eIF1, eIF1A, and eIF3, also bind to the 40S subunit and promote the loading of the TC. The resulting 43S preinitiation complex (PIC) is then loaded onto the 5′ end of an mRNA with the aid of eIF3 and the eIF4 group of factors: the RNA helicase eIF4A; the 5′ 7-methylguanosine cap-binding protein eIF4E; the scaffolding protein eIF4G; and the 40S subunit– and RNA–binding protein eIF4B. Both eIF4A and eIF4E bind to eIF4G and form the eIF4F complex. Once loaded onto the mRNA, the 43S PIC is thought to scan the mRNA in search of an AUG start codon. The process is ATP–dependent and likely requires several RNA helicases, including the DEAD–box protein Ded1p. Recognition of the start site begins with base pairing between the anticodon of tRNAi and the AUG codon. Base pairing then triggers downstream events that commit the PIC to continuing initiation from that point on the mRNA, events that include ejection of eIF1 from its binding site on the 40S subunit, movement of the C-terminal tail (CTT) of eIF1A, and release of phosphate from eIF2, which converts eIF2 to its GDP–bound state. In addition, the initiator tRNA moves from a position that is not fully engaged in the ribosomal P site [termed P(OUT)] to one that is [P(IN)], and the PIC as a whole converts from an open conformation that is conducive to scanning to a closed one that is not. At this stage, eIF2•GDP dissociates from the PIC, and eIF1A and a second GTPase factor, eIF5B, coordinate joining of the large ribosomal subunit to form the 80S initiation complex. In a process that appears to result in conformational reorganization of the complex, eIF5B hydrolyzes GTP and then dissociates along with eIF1A.
The molecular mechanics of eukaryotic translation initiation
In 2020, we made significant progress on our two main projects, despite significant slowdowns owing to the COVID-19 pandemic. We further developed the “Rec-Seq” methodology, which allows transcriptome-wide measurement of translational efficiencies in a fully reconstituted yeast translation initiation system. Unlike the in vivo approach of ribosome profiling, Rec-Seq allows us to isolate events that are directly part of translation initiation and separate them from other connected processes such as transcription, mRNA export from the nucleus, and mRNA decay. In addition, Rec-Seq has an internal control that allows us to make absolute measurements of translation initiation efficiencies rather than only being able to make relative assessments. We can also easily omit factors, change their concentrations, and substitute mutant versions of components, including those that are lethal in vivo, and assess the effects of the changes.
We used the method to probe the functions of several factors that play roles in recruitment of mRNAs to the ribosomal PIC. Our data indicate that the DEAD-box RNA helicase eIF4A is required for efficient recruitment of nearly all cellular mRNAs, whereas Ded1, another DEAD-box RNA helicase, only promotes recruitment of a subset of mRNAs. In particular, mRNAs with long, structured 5′-untranslated regions (5′-UTRs) depend on Ded1 for recruitment to the PIC, whereas mRNAs with short, unstructured 5′-UTRs do not, data that support the idea that Ded1 plays a role in unwinding secondary structures in 5′-UTRs, while eIF4A has a more general role in promoting mRNA loading onto the PIC. We also showed that the factors eIF4B and eIF4G-eIF4E play general stimulatory roles in mRNA recruitment rather than having mRNA specificity.
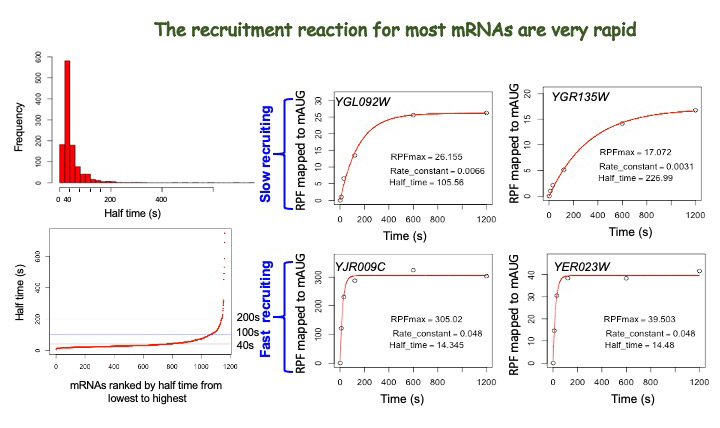
More recently, we expanded the Rec-Seq approach to allow us to measure the kinetics of mRNA recruitment by analyzing several timepoints (Figure 1), an advance that provides us with unprecedented resolution in terms of understanding the influences of different factors and RNA features on the process of mRNA recruitment to the PIC. Using the approach, our initial data demonstrated that the mRNAs that are recruited the fastest to the PIC are those with short, unstructured 5′-UTRs, whereas those that are recruited the slowest have long, structured 5′-UTRs. The data are consistent with the idea that the unwinding of structures in the 5′-UTR is a rate-limiting step in mRNA recruitment to the PIC and in scanning to find the start codon. We anticipate that the ability to measure the kinetics of mRNA recruitment transcriptome-wide in the Rec-Seq system will open many important new avenues for exploration.
Figure 1.
Distribution of rates (half times of reaction) of mRNA recruitment in Rec-Seq experiment (left) and representative time courses for recruitment for individual mRNAs in the same experiment (right)
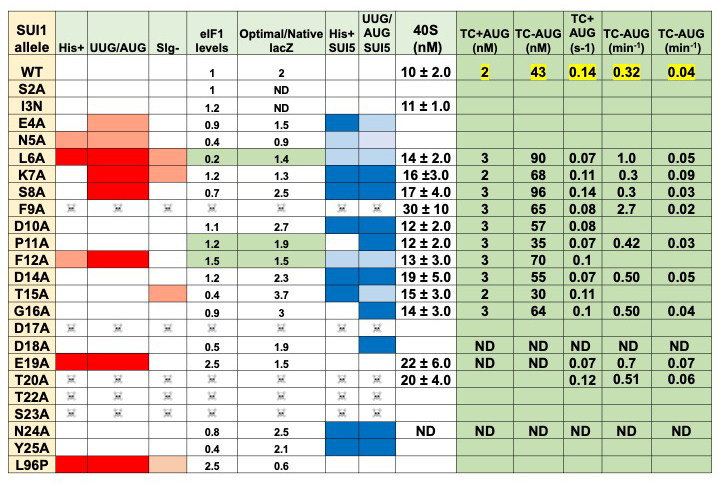
We also made significant progress in our work to elucidate the functions of the unstructured N-terminal tails (NTTs) of the translation initiation factors eIF1 and eIF5. In particular, we completed the initial phenotypic analyses of alanine scanning mutagenesis through the NTT of eIF1. The results reveal many positions at which substitution of the wild-type residue with alanine produces strong phenotypes related to recognition of the mRNA start codon (Figure 2), indicating that the eIF1 NTT plays an important role in start-codon recognition. We expressed and purified key NTT mutants and have begun to study the effects of the mutations on steps in the initiation process in vitro, using the fully reconstituted system. We expect that these studies will shed a great deal of light on the mechanism of action of the eIF1 NTT in start-codon recognition.
Figure 2.
Summary of phenotypes and initial in vitro 43S PIC assembly data for eIF1 NTT mutants
Additional Funding
- Funding is provided via a Memorandum of Understanding (MOU) between NIGMS and NICHD.
Publications
- Zhou F, Zhang H, Kulkarni SD, Lorsch JR, Hinnebusch AG. eIF1 discriminates against suboptimal initiation sites to prevent excessive uORF translation genome-wide. RNA 2020;26:419-438.
- Gulay S, Gupta N, Lorsch JR, Hinnebusch AG. Distinct interactions of eIF4A and eIF4E with RNA helicase Ded1 stimulate translation in vivo. eLife 2020;9:e58243.
Collaborators
- Thomas Dever, PhD, Section on Protein Biosynthesis, NICHD, Bethesda, MD
- Alan Hinnebusch, PhD, Section on Nutrient Control of Gene Expression, NICHD, Bethesda, MD
- Venkatraman Ramakrishnan, PhD, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom
Contact
For more information, email jon.lorsch@nih.gov or visit https://irp.nih.gov/pi/jon-lorsch.