Control of Gene Expression during Development

- Judith A. Kassis, PhD, Head, Section on Gene Expression
- J. Lesley Brown, PhD, Staff Scientist
- Yuzhong Cheng, PhD, Senior Research Technician
- Fountane Chan, BS, Postbaccalaureate Fellow
- Taylor Crawford, BS, Postbaccalaureate Fellow
- Anna Horacek, BS, Postbaccalaureate Fellow
- Joshua Price, BS, Postbaccalaureate Fellow
During development and differentiation, genes become competent to be expressed or are stably silenced in an epigenetically heritable manner. The selective activation/repression of genes leads to differentiation of tissue types. Much evidence supports the model in which modifications of histones in chromatin contribute substantially to determining whether a gene is expressed. Two groups of genes, the Polycomb group (PcG) and Trithorax group (TrxG), are important for inheritance of the silenced and active chromatin state, respectively. In Drosophila, regulatory elements called Polycomb group response elements (PREs) are required for the recruitment of chromatin-modifying PcG protein complexes. TrxG proteins may act through the same or overlapping cis-acting sequences. Our group aims to understand how PcG and TrxG proteins are recruited to DNA. Toward that end, one major project in the lab has been to determine all sequences and DNA–binding proteins required for PRE activity. In the Drosophila genome, there are hundreds of PREs that regulate a similar number of genes, and it was not known whether all PREs are alike. Our recent data showed that there is functional and architectural diversity among PREs, suggesting that PREs adapt to the environment of the gene they regulate. PREs are made up of binding sites for several DNA–binding proteins. Over the years, our lab identified Pho, Pho-like, Spps, and Combgap as DNA proteins that bind to PREs. Our recent genome-wide studies show that different PREs require distinct DNA–binding proteins. In addition, our work illustrates the combinatorial nature and redundancy of PcG recruitment in Drosophila.
A second major project in the lab is to determine how the PREs of the engrailed/invected (en/inv) gene complex control these genes in their native location. Surprisingly, we found that not all PREs are required in vivo, suggesting a redundancy in PRE function. To understand the interplay between PREs and enhancers (sequences important for activation of gene expression), we completed an analysis of the regulatory DNA of the en/inv gene complex. We found that regulatory sequences are spread throughout a region of at least 79kb in that gene complex and that the same enhancers activate both engrailed and invected expression. In addition, we showed that a 79-kb transgene (HA-en79), which contains the en gene and flanking regulatory DNA, is able to rescue a deletion for the entire inv-en locus. Our current studies explore the effects of the chromosomal neighborhood on gene expression. We found that there are subtle differences in the gene expression program of the 79-kb transgene and the endogenous locus. Polycomb domains are flanked by active genes or insulators that limit the size of the domain. We hypothesize that delimiting the size of a Polycomb domain contributes to the stability of both gene activation and repression, making gene expression reproducible and robust.
Polycomb group response elements (PREs)
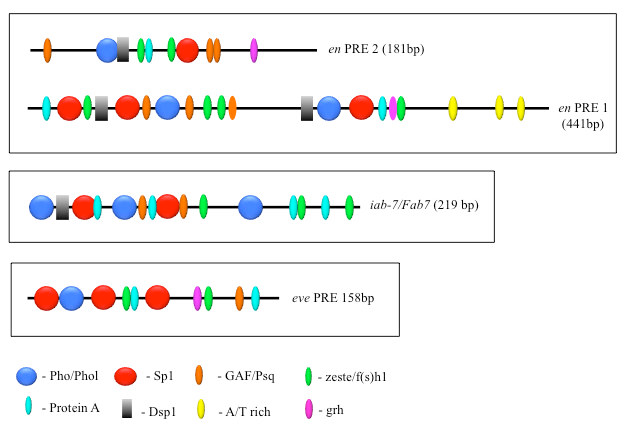
PcG proteins act in protein complexes that repress gene expression by modifying chromatin [Reference 1]. The best-studied PcG protein complexes are PRC1 and PRC2. PRC2 contains the histone methyltransferase Enhancer of Zeste, which tri-methylates lysine 27 on histone H3 (H3K27me3). The chromatin mark H3K27me3 is the signature of PRC2 function. At most well-studied genes, PRC2 acts with PRC1, which binds to H3K27me3 and inhibits chromatin remodeling. In Drosophila, PRC1 and PRC2 are recruited to the DNA by PREs. We are interested in determining how this occurs, and, to that end, we defined all the DNA sequences and are finding all DNA–binding proteins required for the activity of a single 181-bp PRE of the Drosophila engrailed gene (PRE2). We found that binding sites for seven different proteins are required for the activity of the PRE2 (Figure 1). There are several binding sites for some of these proteins. Different PREs have different architectures (Figure 1). Our laboratory identified four PRE DNA–binding proteins: Pho, Phol, Spps, and, most recently, Combgap. The Combgap protein has 10 zinc fingers and recognizes the sequence GTGTGT.
Figure 1. The order, number, and spatial arrangement of consensus-factor binding sites varies in different PREs.
en PRE1 and 2 are from the engrailed gene; iab-7/Fab-7 PRE is from the Abd-B gene; eve PRE is from the even-skipped gene. The symbols represent consensus binding sites for the proteins indicated below. Figure reprinted from Brown JL, Kassis JA. Genetics 2013;195:433.
PRE activity can be studied in transgenes, where a single PRE can recruit PcG protein complexes and silence the expression of a reporter gene. In transgenes, mutation of the Spps, Pho, or Combgap binding sites within the PRE obliterates its ability to recruit PcG proteins, and the reporter gene is expressed. Thus, transcriptional silencing by a single PRE in a transgene requires the combinatorial activity of many DNA–binding proteins. We were interested to determine what happens when one of the PRE DNA–binding proteins (the ‘recruiters’) from the genome is removed, and we examined the effect on PcG recruitment genome-wide. We studied PcG binding genome-wide in mutants that lack the recruiters Spps or Pho [Reference 2]. We found that PcG recruitment to some PREs was completely disrupted, whereas recruitment of PcG proteins was hardly diminished at most PREs. Most PcG domains, which are covered by the chromatin mark H3K27me3, contain several PREs. We believe that the structure of the H3K27me3 domains stabilizes genomic PREs to the loss of one recruiter. However, there are different kinds of PREs, and some are uniquely sensitive to the loss of one recruiter. Our study highlights the complexity and diversity of PcG recruitment mechanisms.
In addition to identifying hundreds of presumptive PREs within Polycomb (H3K27me3) domains, there are thousands of presumptive PREs outside the H3K27me3 domains in larvae [Reference 2]. What is the function of these PREs? Our data suggest that many are tissue-specific PREs, given that H3K27me3 is detected in these locations in some cell types. However, at other locations, no H3K27me3 is present in any cell type. Our on-going studies are addressing the interesting question as to whether these are PREs and to the nature of their function.
The role of PREs at the en gene
The Drosophila engrailed (en) gene encodes a homeodomain protein that plays an important role in the development of many parts of the embryo, including formation of the segments, nervous system, head, and gut. By specifying the posterior compartment of each imaginal disc, en also plays a significant role in the development of the adult. Accordingly, en is expressed in a highly specific and complex manner in the developing organism. The en gene exists in a gene complex with invected (inv), an adjacent gene; inv encodes a protein with a nearly identical homeodomain; en and inv are co-regulated and express proteins with largely redundant functions. Unlike en, inv is dispensable for Drosophila viability in the laboratory.
The en and inv genes exist in a 113kb domain that is covered by the H3K27me3 chromatin mark (Figure 2). Within the en/inv domain there are four major PREs, strong peaks of PcG protein binding. One popular model posits that DNA–binding proteins bound to the PREs recruit PcG protein complexes and that PRC2 tri-methylates histone H3 throughout the domain until PRC2 comes to either an insulator or an actively transcribed gene. There are two PREs upstream of the en transcription unit, PRE1 and PRE2 (Figure 1). Both PREs reside within a 1.5kb fragment located from –1.9kb to –400bp upstream of the major en transcription start site. There are also two major inv PREs, one located at the promoter and another about 6kb upstream of that. Our laboratory showed that all these PREs have the functional properties attributed to PREs in transgenic assays. To test their function at the intact en-inv domain, we set out to delete these PREs from the genome. Given that PREs work as repressive elements, the predicted phenotype of a PRE deletion is a gain-of-function ectopic expression phenotype. Unexpectedly, when we made a 1.5kb deletion removing PRE1 and PRE2, flies were viable and had a partial loss-of-function phenotype in the wing. Similarly, deletion of inv PREs yielded viable flies with no mis-expression of en or inv. Importantly, the H3K27me3 en/inv domain is not disrupted in either of these mutants [Reference 3].
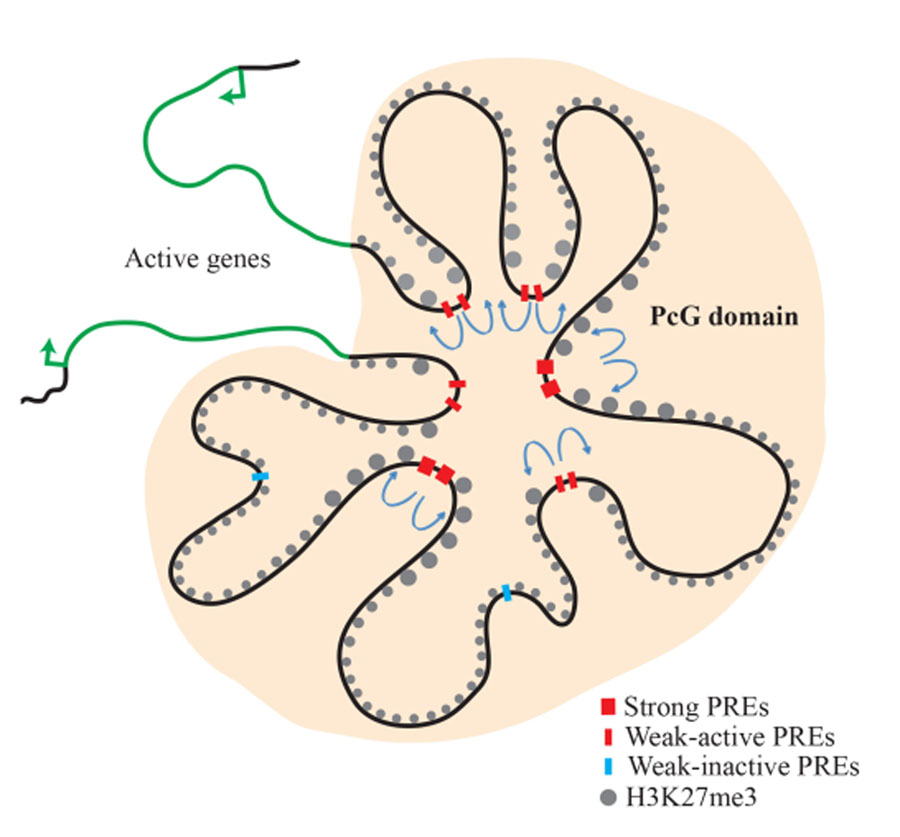
Figure 2. Model of the inv-en PcG domain in the transcriptionally silenced state
The inv and en genes are covered with H3K27me3 and are transcriptionally silent. PcG proteins are associated with this domain. There are strong, constitutive PREs, as well as ‘weak,’ tissue-specific PREs. ‘Weak’ PREs often overlap enhancers and are active in some tissues but inactive in others. Actively transcribed genes remain segregated from the PcG domain and determine the limits of the PcG domain (reprinted from Reference 3).
In Drosophila, PREs are easily recognizable in chromatin immunoprecipitation experiments as discrete peaks of PcG protein binding, but the H3K27me3 mark spreads throughout large regions. PcG proteins are conserved in mammals; however, PcG binding usually does not occur in sharp peaks, and PREs have been much harder to identify. We created a chromosome in which both the en and inv PREs are deleted [Reference 3]. Surprisingly, the flies are viable, and there is no mis-expression of en or inv in embryos or larvae. The question arises as to how PcG proteins are recruited to the inv-en domain in the absence of these PREs. We performed chromatin-immunoprecipitation followed by Next-Gen sequencing (ChIP-seq) on the PcG proteins Pho and Polyhomeotic (Ph). The data showed that, in addition to the large Pho/Ph peaks at the known PREs, there are many smaller Pho/Ph peaks within the inv/en domain. We found that those peaks may also function as PREs. Thus, rather than a few PREs, there are many PREs controlling inv-en expression, and some may act in tissue-specific ways [Reference 3]. Our work shows that there are two types of PREs in Drosophila: strong, constitutive PREs and tissue-specific PREs that tend to overlap with enhancers (Figure 2).
The inv-en gene complex is flanked by tou and E(Pc), two ubiquitously expressed genes (Figures 2 & 3). The H3K27me3 mark stops at the two genes. We believe that it is the transcription of these two genes that blocks the spreading of the H3K27me3 mark and stabilizes the repression of inv and en by PcG proteins. To test this assumption, we made a large transgene marked by HA–tagged Engrailed protein. A 79-kb HA-en transgene was able to correctly express En and completely rescue inv-en double mutants. We inserted the transgene into other places in the Drosophila genome [Reference 4]. Our data showed that, while the information to form the H3K27me3 domain is contained within the 79-kb HA-en transgene, the structure of the H3K27me3 domains differs from that at the endogenous locus. Specifically, the H3K27me3 mark spread beyond the transgene into flanking DNA. Further, enhancers within the 79-kb HA-en transgene could interact with some flanking genes and drive their expression in subsets of the En pattern. Also, removal of the PREs from the transgene led to loss of PcG silencing in the abdominal segments of the flies. These data provide evidence that the endogenous inv-en domain imparts stability to the locus and facilitates both transcriptional activation and silencing of these two developmentally important genes.
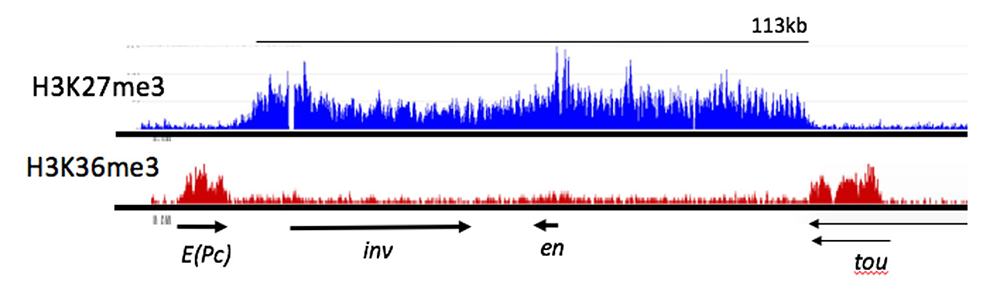
Figure 3. ChIP-seq profile showing H3K27me3 and H3K36me3 binding to the e(Pc), inv, en, and tou genes in Drosophila
H3K27me3, a mark deposited by PcG protein complex PRC2, is bound from the 3′ end of the tou gene to the 3′ end of the E(Pc) gene. Arrows indicate the direction and extent of the transcription units for the genes shown. H3K36me3 is a mark of actively transcribed genes and is bound to E(Pc) and tou. Samples from Drosophila 3rd instar larvae, brains, and discs. In these tissues, at least 80% of the cells do not express inv or en. Data from Reference 4.
Enhancer-promoter communication
Enhancers are often located tens or even hundreds of kb away from their promoter, sometimes even closer to the promoters of genes other than the one they activate. Several years ago, we showed that en enhancers can act over large distances, even skipping over other transcription units, choosing the en promoter over those of neighboring genes. Such specificity is achieved in at least three ways. First, early-acting enhancers that drive engrailed expression in stripes exhibit promoter specificity. Second, a proximal promoter-tethering element is required for the action of the imaginal disc enhancer(s). Our data point to two partially redundant promoter-tethering elements. Third, the long-distance action of en enhancers requires a combination of the en promoter and sequences within or closely linked to the promoter-proximal PREs. The data show that several mechanisms ensure proper enhancer-promoter specificity at the Drosophila en locus, providing one of the first detailed views of how promoter-enhancer specificity is achieved.
As a follow-up to these studies, we located all the enhancers that regulate the transcription of en and the closely linked co-regulated inv gene (Figure 4). Our dissection of inv-en–regulatory DNA showed that most enhancers are spread throughout a 62kb region. We used two types of constructs to analyze the function of this DNA: P-element–based reporter constructs with small pieces of DNA fused to the en promoter driving lacZ expression (Figure 4); and large constructs with HA–tagged en and inv inserted in the genome with the phiC31 integrase. In addition, we generated deletions of inv and en DNA in situ and assayed their effects on inv/en expression. Our results support and extend our knowledge of inv-en regulation. First, inv and en share regulatory DNA, most of which flanks the en transcription unit. In support of this finding, a 79-kb HA-en transgene can rescue inv en double mutants into viable, fertile adults. In contrast, an 84-kb HA-inv transgene lacks most of the enhancers for inv and en expression. Second, there are several enhancers for inv/en stripes in embryos; some may be redundant, but others play discrete roles at different stages of embryonic development. Finally, no small reporter construct gave expression in the posterior compartment of imaginal discs, a hallmark of inv/en expression. Robust expression of HA-en in the posterior compartment of imaginal discs is evident from the 79-kb HA-en transgene, while a 45-kb HA-en transgene gives weaker, variable imaginal disc expression. We suggest that the activity of the imaginal disc enhancer(s) depends on the chromatin structure of the inv-en domain. We are currently investigating the properties of the inv/en imaginal disc enhancer(s) using a variety of methods, including deleting them from the endogenous inv/en domain using CRISPR/cas9.
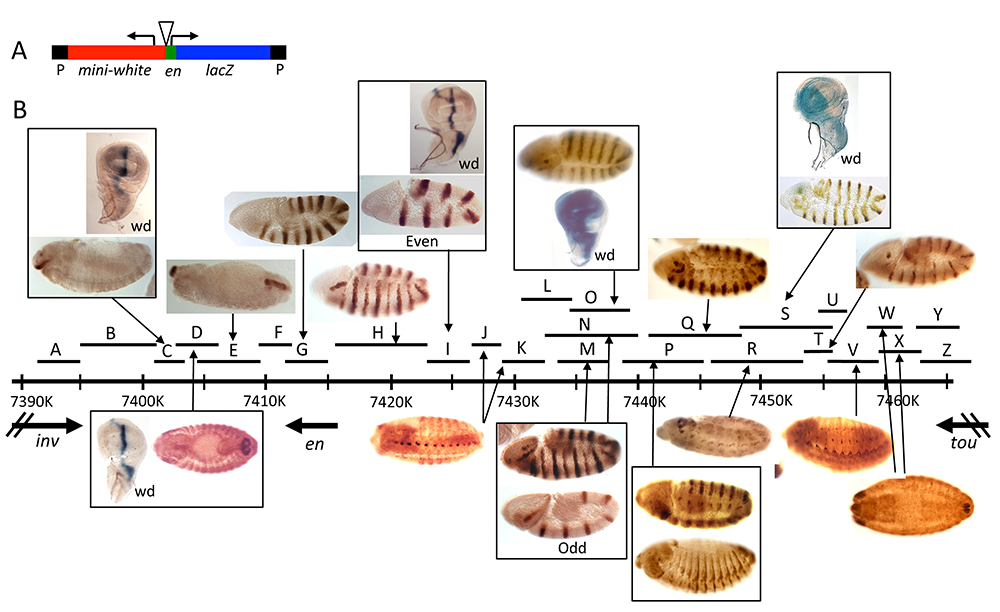
Figure 4. Enhancers of the invected and engrailed genes
A. P-element vector (P[en]), used to assay the function of en–regulatory DNA, contains the en promoter, 396bp of upstream sequences, and an untranslated leader fusion between the en transcript and the Adh-lacZ reporter gene. inv/en DNA fragments were added to this vector at the location of the triangle.
B. The extent of each fragment cloned into P[en] is shown as a black line with a letter above the inv/en genomic DNA map (indicated by a long black line with hatch marks at 10kb intervals; numbers are coordinates on chromosome 2R, genome release v5). Expression pattern in embryos or the wing imaginal disc (wd) are shown above or below the genomic DNA, with arrows pointing to the fragment(s) that generate(s) the pattern. Figure reprinted from Cheng Y et al. Dev Biol 2014;395:131.
Defining the ends of Polycomb domains in Drosophila
Actively transcribed genes flank many Polycomb domains, and previous genomic studies showed that inhibition of transcription using chemical inhibitors leads to a spreading of H3K27me3 in the genome. We conducted a genome-wide analysis of Polycomb boundaries in Drosophila larvae [Reference 5]. We found six different types of Polycomb-domain boundaries, including those made by insulator proteins, and actively transcribed genes. The inv-en Polycomb domain is flanked by two actively transcribed genes, E(Pc) and tou (Figure 3). Insertion of a transcriptional stop within the tou gene causes an extension of the H3K27me3 mark to the point of active transcription. We also suggest that active transcription limits the range of inv-en enhancers and that promoter specificity is important for inv-en enhancer activity [Reference 5]. Currently, we are working to add actively transcribed genes, as well as, separately, to add insulators to delimit the ends of the 79-kb transgene. We hypothesize that adding boundaries will increase the accuracy and robustness of the expression of the transgene.
Publications
- Kuroda MI, Kang H, De S, Kassis JA. Dynamic competition of Polycomb and Trithorax in transcriptional programming. Ann Rev Biochem 2020;89:235-253.
- Brown JL, Sun M, Kassis JA. Global changes of H3K27me3 domains and Polycomb group protein distribution in the absence of recruiters Spps or Pho. Proc Natl Acad Sci USA 2018;E1839-E1848.
- De S, Mitra A, Cheng A, Pfeifer Y, Kassis JA. Formation of a Polycomb-domain in the absence of strong Polycomb Response Elements. PLoS Genetics 2016;12:e1006200.
- De S, Cheng Y, Sun M, Gehred ND, Kassis JA. Structure and function of an ectopic Polycomb chromatin domain. Sci Adv 2019;5:eaau9739.
- De S, Gehred ND, Fujioka M, Chan FW, Jaynes JB, Kassis, JA. Defining the boundaries of Polycomb domains in Drosophila. Genetics 2020;216(3):689-700.
Collaborators
- Karl Pfeifer, PhD, Section on Epigenetics, NICHD, Bethesda, MD
Contact
For more information, email jkassis@mail.nih.gov or visit http://kassislab.nichd.nih.gov.